Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE G02374QK
G02374QK
Summary
- GlyTouCan ID
-
G02374QK
- Subsumption Level
- Fully-defined saccharide
3D Structures
- GlycoShape
- G02374QK
Organisms
| Organisms | Evidence |
|---|---|
| Pseudomonas syringae | |
| Pseudomonas syringae pv. delphinii |
Taxonomic Hierarchy
Sequence Descriptors
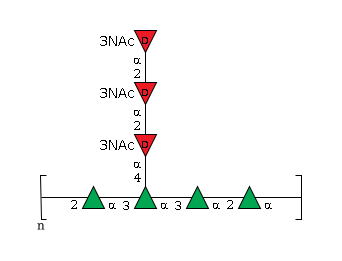
- GlycoCT
-
RES 1r:r1 REP REP1:5o(2+1)2d=-1--1 RES 2b:a-lman-HEX-1:5|6:d 3b:a-lman-HEX-1:5|6:d 4b:a-lman-HEX-1:5|6:d 5b:a-lman-HEX-1:5|6:d 6b:a-dgal-HEX-1:5|6:d 7b:a-dgal-HEX-1:5|6:d 8b:a-dgal-HEX-1:5|6:d 9s:n-acetyl 10s:n-acetyl 11s:n-acetyl LIN 1:2o(2+1)3d 2:3o(3+1)4d 3:4o(3+1)5d 4:4o(4+1)6d 5:6o(2+1)7d 6:7o(2+1)8d 7:8d(3+1)9n 8:7d(3+1)10n 9:6d(3+1)11n
- WURCS
- WURCS=2.0/2,7,7/[a2211m-1a_1-5][a2112m-1a_1-5_3*NCC/3=O]/1-1-1-1-2-2-2/a2-b1_b3-c1_c3-d1_c4-e1_e2-f1_f2-g1_a1-d2~n
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 12059776 | Structure of the O-polysaccharide of Pseudomonas syringae pv. delphinii NCPPB 1879(T) having side chains of 3-acetamido-3,6-dideoxy-D-galactose residues | Zdorovenko EL | 2002 May |
|