Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
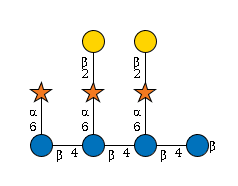
MIRAGE G06184LI
G06184LI
Summary
- GlyTouCan ID
-
G06184LI
- IUPAC Condensed
- Xyl(a1-6)Glc(b1-4)[Gal(b1-2)Xyl(a1-6)]Glc(b1-4)[Gal(b1-2)Xyl(a1-6)]Glc(b1-4)Glc(b1-
- Subsumption Level
- Fully-defined saccharide
- Monoisotopic Mass
- 1386.45
3D Structures
GlycoShape
Organisms
| Organisms | Evidence |
|---|---|
| Tamarindus indica |
Taxonomic Hierarchy
Sequence Descriptors
- GlycoCT
-
RES 1b:b-dglc-HEX-1:5 2b:b-dglc-HEX-1:5 3b:b-dglc-HEX-1:5 4b:b-dglc-HEX-1:5 5b:a-dxyl-PEN-1:5 6b:a-dxyl-PEN-1:5 7b:b-dgal-HEX-1:5 8b:a-dxyl-PEN-1:5 9b:b-dgal-HEX-1:5 LIN 1:1o(4+1)2d 2:2o(4+1)3d 3:3o(4+1)4d 4:4o(6+1)5d 5:3o(6+1)6d 6:6o(2+1)7d 7:2o(6+1)8d 8:8o(2+1)9d
- WURCS
- WURCS=2.0/3,9,8/[a2122h-1b_1-5][a212h-1a_1-5][a2112h-1b_1-5]/1-1-1-1-2-2-3-2-3/a4-b1_b4-c1_b6-h1_c4-d1_c6-f1_d6-e1_f2-g1_h2-i1
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 31324716 | Substrate specificity, regiospecificity, and processivity in glycoside hydrolase family 74 | Arnal G | 2019 Sep 06 |
|
| 27637560 | Structure of Arabidopsis thaliana FUT1 Reveals a Variant of the GT-B Class Fold and Provides Insight into Xyloglucan Fucosylation | Rocha J | 2016 Sep 16 |
|
| 26929175 | Functional and structural characterization of a potent <scp>GH</scp>74 endo‐xyloglucanase from the soil saprophyte Cellvibrio japonicus unravels the first step of xyloglucan degradation | Attia M | 2016 Mar 30 |
|
| 16772298 | Crystal Structures of Clostridium thermocellum Xyloglucanase, XGH74A, Reveal the Structural Basis for Xyloglucan Recognition and Degradation | Martinez-Fleites C | 2006 May 22 |
|