Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
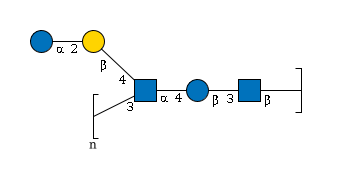
MIRAGE G10235UA
G10235UA
Summary
- GlyTouCan ID
-
G10235UA
- Subsumption Level
- Fully-defined saccharide
Organisms
| Organisms | Evidence |
|---|---|
| Proteus mirabilis |
Sequence Descriptors
- GlycoCT
-
RES 1r:r1 REP REP1:5o(3+1)2d=-1--1 RES 2b:b-dglc-HEX-1:5 3s:n-acetyl 4b:b-dglc-HEX-1:5 5b:a-dglc-HEX-1:5 6s:n-acetyl 7b:b-dgal-HEX-1:5 8b:a-dglc-HEX-1:5 LIN 1:2d(2+1)3n 2:2o(3+1)4d 3:4o(4+1)5d 4:5d(2+1)6n 5:5o(4+1)7d 6:7o(2+1)8d
- WURCS
- WURCS=2.0/5,5,5/[a2122h-1b_1-5_2*NCC/3=O][a2122h-1b_1-5][a2122h-1a_1-5_2*NCC/3=O][a2112h-1b_1-5][a2122h-1a_1-5]/1-2-3-4-5/a3-b1_b4-c1_c4-d1_d2-e1_a1-c3~n
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 21999547 | Serotyping of Proteus mirabilis clinical strains based on lipopolysaccharide O-polysaccharide and core oligosaccharide structures | Kaca W | 2011 Jul |
|
| 20305038 | Structure and serology of O-antigens as the basis for classification of Proteus strains | Knirel YA | 2011 Feb |
|
| 20618127 | Structural analysis of lipopolysaccharides from Gram-negative bacteria | Kabanov DS | 2010 Apr |
|
| 15013399 | Structure of the neutral O-polysaccharide and biological activities of the lipopolysaccharide of Proteus mirabilis O20 | Kondakova AN | 2004 Feb 25 |
|