Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE G22768VO
G22768VO
Summary
- GlyTouCan ID
-
G22768VO
- IUPAC Condensed
- Man(a1-3)[Man(a1-6)]Man(b1-4)GlcNAc(b1-4)GlcNAc(b1-
- Motifs
- N-Glycan core basic
- Subsumption Level
- Fully-defined saccharide
- Monoisotopic Mass
- 910.33
External Links
- GlyConnect structure
- GlyGen
- ChEBI
- PubChem Compound
- PubChem Substance
- CarbBank
3D Structures
Organisms
| Organisms | Evidence |
|---|---|
| Mus musculus (house mouse) | |
| Bos taurus (domestic cattle) | |
| Rana | |
| Rattus norvegicus (Norway rat) | |
| Severe acute respiratory syndrome coronavirus 2 |
Taxonomic Hierarchy
GlycoGene Database (GGDB)
GALAXY
Core Protein
| UniProt ID | Protein Name | Reference | Source |
|---|---|---|---|
| Q40379 | Ribonuclease S-6 | ||
| Q7Z1F8 | Arylphorin |
Sequence Descriptors
- GlycoCT
-
RES 1b:b-dglc-HEX-1:5 2s:n-acetyl 3b:b-dglc-HEX-1:5 4s:n-acetyl 5b:b-dman-HEX-1:5 6b:a-dman-HEX-1:5 7b:a-dman-HEX-1:5 LIN 1:1d(2+1)2n 2:1o(4+1)3d 3:3d(2+1)4n 4:3o(4+1)5d 5:5o(3+1)6d 6:5o(6+1)7d
- WURCS
- WURCS=2.0/3,5,4/[a2122h-1b_1-5_2*NCC/3=O][a1122h-1b_1-5][a1122h-1a_1-5]/1-1-2-3-3/a4-b1_b4-c1_c3-d1_c6-e1
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 22802656 | Crystal structure of IgE bound to its B-cell receptor CD23 reveals a mechanism of reciprocal allosteric inhibition with high affinity receptor FcεRI | Dhaliwal B | 2012 Jul 16 |
|
| 20356844 | Structural Basis for Substrate Selectivity in Human Maltase-Glucoamylase and Sucrase-Isomaltase N-terminal Domains | Sim L | 2010 Jun 04 |
|
| 18446213 | Correction: Intervening with Urinary Tract Infections Using Anti-Adhesives Based on the Crystal Structure of the FimH–Oligomannose-3 Complex | Wellens A | 2008 Jun 09 |
|
| 18615077 | Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor | Lee JE | 2008 Jul 10 |
|
| 18272523 | Structural Insights into the Mechanism of pH-dependent Ligand Binding and Release by the Cation-dependent Mannose 6-Phosphate Receptor | Olson LJ | 2008 Apr 11 |
|
| 17264077 | Dynamic Developmental Elaboration of N-Linked Glycan Complexity in the Drosophila melanogaster Embryo | Aoki K | 2007 Mar 23 |
|
| 16897177 | Glycomic studies of Drosophila melanogaster embryos | North SJ | 2006 Jul |
|
| 16339150 | The Drosophila fused lobes gene encodes an N-acetylglucosaminidase involved in N-glycan processing | Leonard R | 2006 Feb 24 |
|
| 15937331 | Crystal structures of the HIV-1 inhibitory cyanobacterial protein MVL free and bound to Man3GlcNAc2: structural basis for specificity and high-affinity binding to the core pentasaccharide from n-linked oligomannoside | Williams DC | 2005 Aug 12 |
|
| 15620693 | A novel β1,3‐N‐acetylglucosaminyltransferase (β3Gn‐T8), which synthesizes poly‐N‐acetyllactosamine, is dramatically upregulated in colon cancer | Ishida H | 2004 Nov 28 |
|
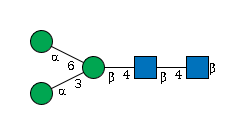
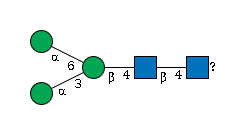 G00026MO
G00026MO
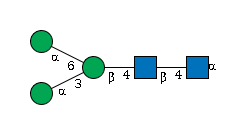 G43547MI
G43547MI
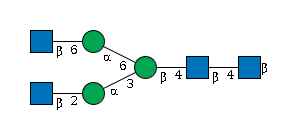 G74090KZ
G74090KZ
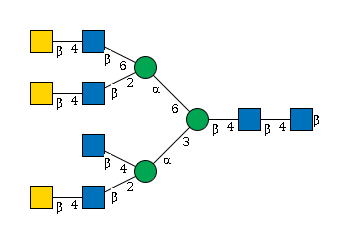 G74438AB
G74438AB
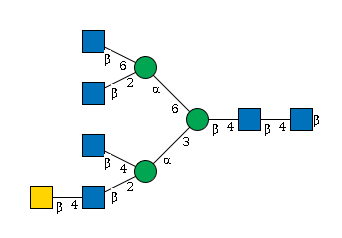 G77639OD
G77639OD
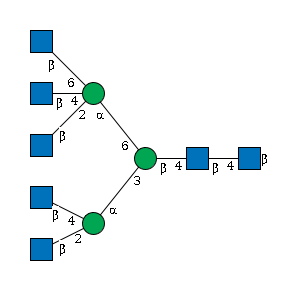 G77974XY
G77974XY
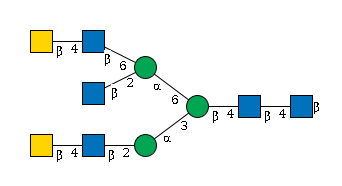 G81923ER
G81923ER
