Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE G49108TO
G49108TO
Summary
- GlyTouCan ID
-
G49108TO
- IUPAC Condensed
- GlcNAc(b1-
External Links
- GlyConnect structure
- GlyGen
- ChEBI
- PubChem Compound
- PubChem Substance
- The Human Metabolome Database
- CarbBank
-
- 9
- 3780
- 3968
- 4708
- 25653
- 28493
- 29021
- 29527
- 29549
- 31775
- 32028
- 32903
- 33539
- 36048
- 36270
- 37035
- 37525
- 37675
- 37677
- 37678
- 39452
- 39454
- 39456
- 39609
- 39611
- 39613
- 39614
- 40149
- 40150
- 40151
- 40355
- 40591
- 40600
- 42059
- 42207
- 42220
- 43184
- 43893
- 43925
- 43926
- 43931
- 43932
- 43941
- 44395
- 44396
- 44397
- 44549
- 46453
- 47268
- 47468
- 47537
- 47603
- 47796
- 48630
- 48812
- 48813
- 49006
- 49732
- 50589
- 50736
3D Structures
- GLYCAM
- DGlcpNAcb1-OH
Organisms
| Organisms | Evidence |
|---|---|
| Mus musculus (house mouse) | |
| Rattus norvegicus (Norway rat) | |
| Homo sapiens (human) | |
| Drosophila melanogaster (fruit fly) | |
| Streptococcus pneumoniae |
Taxonomic Hierarchy
GlycoGene Database (GGDB)
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| B3GALT1 | UDP-Gal |
 G49108TO
G49108TO
|
Lemieux |
 G00056MO
G00056MO
|
Lemieux | |
| B4GALNT3 | UDP-GalNAc |
 G49108TO
G49108TO
|
R |
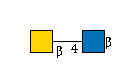 G00057MO
G00057MO
|
R | |
| B3GALNT2 | UDP-GalNAc |
 G49108TO
G49108TO
|
R |
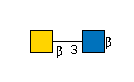 G36387CV
G36387CV
|
R | |
| B4GALNT4 | UDP-GalNAc |
 G49108TO
G49108TO
|
R |
 G00057MO
G00057MO
|
R | |
| B4GALT1 | UDP-Glc |
 G49108TO
G49108TO
|
R |
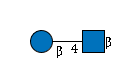 G38884XS
G38884XS
|
R |
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| B4GALT5 | (not applicable) |
 G49108TO
G49108TO
|
[beta]-S-pNP |
 G00055MO
G00055MO
|
[beta]-S-pNP | |
| B4GALT3 | UDP-Gal |
 G49108TO
G49108TO
|
Benzyl-[beta] |
 G00055MO
G00055MO
|
Benzyl-[beta] | |
| B4GALT4 | UDP-Gal |
 G49108TO
G49108TO
|
[beta]-1-Benzl |
 G00055MO
G00055MO
|
[beta]-1-Benzl | |
| B3GALT2 | UDP-Gal |
 G49108TO
G49108TO
|
Lemieux |
 G00056MO
G00056MO
|
Lemieux | |
| B4GALT4 | UDP-Gal |
 G49108TO
G49108TO
|
[beta]-1-4-methyl-umbelliferyl |
 G00055MO
G00055MO
|
[beta]-1-4-methyl-umbelliferyl |
Core Protein
| UniProt ID | Protein Name | Reference | Source |
|---|---|---|---|
| Q9HC96 | Calpain-10 | ||
| Q9HCB6 | Spondin-1 | ||
| Q9HCD5 | Nuclear receptor coactivator 5 | ||
| Q9HCD6 | Protein TANC2 | ||
| Q9HCE3 | Zinc finger protein 532 | ||
| Q9HCF6 | Transient receptor potential cation channel subfamily M member 3 | ||
| Q9HCH5 | Synaptotagmin-like protein 2 | ||
| Q9HCJ0 | Trinucleotide repeat-containing gene 6C protein | ||
| Q9HCK1 | DBF4-type zinc finger-containing protein 2 | ||
| Q9HCK8 | Chromodomain-helicase-DNA-binding protein 8 |
GlycoEpitope
- Epitope ID
- EP0004
Pathway
| Pathway Name | Organism |
|---|---|
| Antimicrobial peptides | Xenopus tropicalis |
| Antimicrobial peptides | Rattus norvegicus |
| Antimicrobial peptides | Danio rerio |
| Antimicrobial peptides | Bos taurus |
| Antimicrobial peptides | Canis familiaris |
| Antimicrobial peptides | Sus scrofa |
| Antimicrobial peptides | Dictyostelium discoideum |
| Antimicrobial peptides | Mus musculus |
| Antimicrobial peptides | Drosophila melanogaster |
| Antimicrobial peptides | Homo sapiens |
Sequence Descriptors
- GlycoCT
-
RES 1b:b-dglc-HEX-1:5 2s:n-acetyl LIN 1:1d(2+1)2n
- WURCS
- WURCS=2.0/1,1,0/[a2122h-1b_1-5_2*NCC/3=O]/1/
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 38129107 | Loss of NDST1 N-sulfotransferase activity is associated with autosomal recessive intellectual disability | Khosrowabadi E | 2023 Dec 21 |
|
| 38059672 | Structural and time‐resolved mechanistic investigations of protein hydrolysis by the acidic proline‐specific endoprotease from Aspergillus niger | Pijning T | 2023 Dec 20 |
|
| 38118176 | Molecular Basis for Inhibition of Heparanases and β-Glucuronidases by Siastatin B | Chen Y | 2023 Dec 20 |
|
| 38139429 | ER Stress-Perturbed Intracellular Protein O-GlcNAcylation Aggravates Podocyte Injury in Diabetes Nephropathy | Song S | 2023 Dec 18 |
|
| 38109549 | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2 | Rao X | 2023 Dec 18 |
|
| 37989312 | Broad receptor tropism and immunogenicity of a clade 3 sarbecovirus | Lee J | 2023 Dec 13 |
|
| 38085789 | Discovery of High Affinity Cyclic Peptide Ligands for Human ACE2 with SARS-CoV-2 Entry Inhibitory Activity | Bedding MJ | 2023 Dec 12 |
|
| 38083975 | T-2 Toxin-Mediated β-Arrestin-1 O-GlcNAcylation Exacerbates Glomerular Podocyte Injury via Regulating Histone Acetylation | Li T | 2023 Dec 11 |
|
| 38054441 | O-GlcNAcPRED-DL: Prediction of Protein O-GlcNAcylation Sites Based on an Ensemble Model of Deep Learning | Hu F | 2023 Dec 06 |
|
| 38047498 | Top/Middle-Down Characterization of α-Synuclein Glycoforms | Miller SA | 2023 Dec 04 |
|
 G14843DJ
G14843DJ
 G68544GH
G68544GH
