Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE G71142DF
G71142DF
Summary
- GlyTouCan ID
-
G71142DF
- IUPAC Condensed
- Glc(b1-
- Motifs
- Glucosylceramide
External Links
- GlyGen
- ChEBI
- PubChem Compound
- PubChem Substance
- The Human Metabolome Database
3D Structures
- GlycoNAVI
- G71142DF
- GLYCAM
- DGlcpb1-OH
Organisms
| Organisms | Evidence |
|---|---|
| Mus musculus (house mouse) | |
| Clostridium | |
| Chloroflexus aurantiacus | |
| Homo sapiens (human) | |
| Glycine max (soybean) |
Taxonomic Hierarchy
GlycoGene Database (GGDB)
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| B4GALT5 | (not applicable) |
 G71142DF
G71142DF
|
Ceramide |
 G84224TW
G84224TW
|
Ceramide | |
| B4GALT6 | UDP-Gal |
 G71142DF
G71142DF
|
Cer |
 G84224TW
G84224TW
|
Cer | |
| GXYLT1 | UDP-Xyl |
 G71142DF
G71142DF
|
Ser |
 G01629YL
G01629YL
|
Ser | |
| GXYLT2 | UDP-Xyl |
 G71142DF
G71142DF
|
Ser |
 G01629YL
G01629YL
|
Ser |
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| B3GALT1 | UDP-Gal |
 G71142DF
G71142DF
|
Cer |
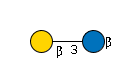 G31761VE
G31761VE
|
Cer | |
| B4GALT3 | UDP-Gal |
 G71142DF
G71142DF
|
[beta]1-Cer |
 G84224TW
G84224TW
|
[beta]1-Cer | |
| GXYLT1 | UDP-Xyl |
 G71142DF
G71142DF
|
Ser |
 G01629YL
G01629YL
|
Ser | |
| GXYLT2 | UDP-Xyl |
 G71142DF
G71142DF
|
Ser |
 G01629YL
G01629YL
|
Ser |
KEGG BRITE Database
- KEGG ID
- Enzyme
- Reactions
Pathway
| Pathway Name | Organism |
|---|---|
| E.coli O102 | Escherichia coli |
| E.coli O103 | Escherichia coli |
| E.coli O130 | Escherichia coli |
| E.coli O134 | Escherichia coli |
| E.coli O137 | Escherichia coli |
| E.coli O139 | Escherichia coli |
| E.coli O141 | Escherichia coli |
| E.coli O150 | Escherichia coli |
| E.coli O152 | Escherichia coli |
| E.coli O157 | Escherichia coli |
Sequence Descriptors
- GlycoCT
-
RES 1b:b-dglc-HEX-1:5
- WURCS
- WURCS=2.0/1,1,0/[a2122h-1b_1-5]/1/
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 35698170 | Lignocellulose degradation in Protaetia brevitarsis larvae digestive tract: refining on a tightly designed microbial fermentation production line | Wang K | 2022 Jun 13 |
|
| 35687305 | Asymmetric Hydrogenation of C = C Bonds in a SpinChem Reactor by Immobilized Old Yellow Enzyme and Glucose Dehydrogenase | Ma T | 2022 Jun 10 |
|
| 35661053 | Characterisation of recombinant GH 3 β-glucosidase from β-glucan producing Levilactobacillus brevis TMW 1.2112 | Bockwoldt JA | 2022 Jun 04 |
|
| 35655182 | Construction and characterization of BsGDH-CatIB variants and application as robust and highly active redox cofactor regeneration module for biocatalysis | Küsters K | 2022 Jun 02 |
|
| 35653012 | Metabolomic Profiling of Phloem Sap from Different Pine Species and Implications on Black Capuchin | Silva LMA | 2022 Jun 02 |
|
| 35869468 | Metabolomic biomarkers related to non-suicidal self-injury in patients with bipolar disorder | Guo X | 2022 Jul 22 |
|
| 35838870 | Insights into the development of pentylenetetrazole-induced epileptic seizures from dynamic metabolomic changes | Zhao X | 2022 Jul 15 |
|
| 35833950 | Biochemical characterization of a novel glucose-tolerant GH3 β-glucosidase (Bgl1973) from Leifsonia sp. ZF2019 | He Y | 2022 Jul 14 |
|
| 35834078 | Carboxymethyl cellulose nanocolloids anchored Pd(0) nanoparticles (CMC@Pd NPs): synthesis, characterization, and catalytic application in transfer hydrogenation | Mekkaoui AA | 2022 Jul 14 |
|
| 35819689 | NMR-Based Metabolomic Profiling of Mungbean Infected with Mungbean Yellow Mosaic India Virus | Maravi DK | 2022 Jul 12 |
|
 G80753ZP
G80753ZP
 G58161NS
G58161NS
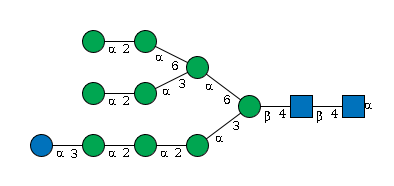 G24620AI
G24620AI
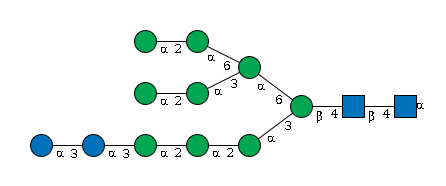 G98256YA
G98256YA
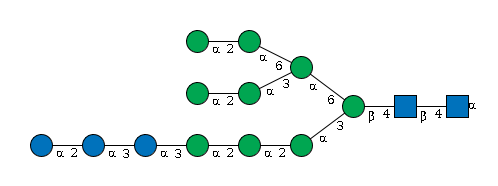 G72079JG
G72079JG
