Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE G71142DF
G71142DF
Summary
- GlyTouCan ID
-
G71142DF
- IUPAC Condensed
- Glc(b1-
- Motifs
- Glucosylceramide
External Links
- GlyGen
- ChEBI
- PubChem Compound
- PubChem Substance
- The Human Metabolome Database
3D Structures
- GlycoNAVI
- G71142DF
- GLYCAM
- DGlcpb1-OH
Organisms
| Organisms | Evidence |
|---|---|
| Mus musculus (house mouse) | |
| Clostridium | |
| Chloroflexus aurantiacus | |
| Homo sapiens (human) | |
| Glycine max (soybean) |
Taxonomic Hierarchy
GlycoGene Database (GGDB)
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| B4GALT5 | (not applicable) |
 G71142DF
G71142DF
|
Ceramide |
 G84224TW
G84224TW
|
Ceramide | |
| B4GALT6 | UDP-Gal |
 G71142DF
G71142DF
|
Cer |
 G84224TW
G84224TW
|
Cer | |
| GXYLT1 | UDP-Xyl |
 G71142DF
G71142DF
|
Ser |
 G01629YL
G01629YL
|
Ser | |
| GXYLT2 | UDP-Xyl |
 G71142DF
G71142DF
|
Ser |
 G01629YL
G01629YL
|
Ser |
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| B3GALT1 | UDP-Gal |
 G71142DF
G71142DF
|
Cer |
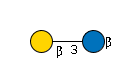 G31761VE
G31761VE
|
Cer | |
| B4GALT3 | UDP-Gal |
 G71142DF
G71142DF
|
[beta]1-Cer |
 G84224TW
G84224TW
|
[beta]1-Cer | |
| GXYLT1 | UDP-Xyl |
 G71142DF
G71142DF
|
Ser |
 G01629YL
G01629YL
|
Ser | |
| GXYLT2 | UDP-Xyl |
 G71142DF
G71142DF
|
Ser |
 G01629YL
G01629YL
|
Ser |
KEGG BRITE Database
- KEGG ID
- Enzyme
- Reactions
Pathway
| Pathway Name | Organism |
|---|---|
| E.coli O160 | Escherichia coli |
| E.coli O163 | Escherichia coli |
| E.coli O164 | Escherichia coli |
| E.coli O165 | Escherichia coli |
| E.coli O166 | Escherichia coli |
| E.coli O169 | Escherichia coli |
| E.coli O171 | Escherichia coli |
| E.coli O173 | Escherichia coli |
| E.coli O18B | Escherichia coli |
| E.coli O18B1 | Escherichia coli |
Sequence Descriptors
- GlycoCT
-
RES 1b:b-dglc-HEX-1:5
- WURCS
- WURCS=2.0/1,1,0/[a2122h-1b_1-5]/1/
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 37247149 | Metabolic variation and oxidative stress response of blue mussels (Mytilus sp.) perturbed by norfloxacin exposure | Zhou X | 2023 May 29 |
|
| 37231524 | Abnormal glycosylation in glioma: related changes in biology, biomarkers and targeted therapy | Yue J | 2023 May 26 |
|
| 37149838 | Deciphering host–pathogen interaction during Streptomyces spp. infestation of potato | Haq IU | 2023 May 07 |
|
| 37002508 | Integerrima A–E, phenylethanoid glycosides from the stem of Callicarpa integerrima | Jiang W | 2023 Mar 31 |
|
| 36959350 | Complete genome of Sphingomonas paucimobilis ZJSH1, an endophytic bacterium from Dendrobium officinale with stress resistance and growth promotion potential | Li J | 2023 Mar 23 |
|
| 36854658 | Correction: Dying transplanted neural stem cells mediate survival bystander effects in the injured brain | Han W | 2023 Mar 01 |
|
| 37781051 | Enzymatic time−temperature indicator with cysteine-loaded chitosan microspheres/silver nanoparticles | Cho HW | 2023 Jun 26 |
|
| 37335345 | Acidobacteria members harbour an abundant and diverse carbohydrate-active enzymes (cazyme) and secreted proteasome repertoire, key factors for potential efficient biomass degradation | Coluccia M | 2023 Jun 19 |
|
| 37308810 | Potential pathways and genes expressed in Chrysanthemum in response to early fusarium oxysporum infection | Miao W | 2023 Jun 13 |
|
| 37289241 | Valorization of whey-based side streams for microbial biomass, molecular hydrogen, and hydrogenase production | Poladyan A | 2023 Jun 08 |
|
 G80753ZP
G80753ZP
 G58161NS
G58161NS
 G24620AI
G24620AI
 G98256YA
G98256YA
 G72079JG
G72079JG
