Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE G71142DF
G71142DF
Summary
- GlyTouCan ID
-
G71142DF
- IUPAC Condensed
- Glc(b1-
- Motifs
- Glucosylceramide
External Links
- GlyGen
- ChEBI
- PubChem Compound
- PubChem Substance
- The Human Metabolome Database
3D Structures
- GlycoNAVI
- G71142DF
- GLYCAM
- DGlcpb1-OH
Organisms
| Organisms | Evidence |
|---|---|
| Mus musculus (house mouse) | |
| Clostridium | |
| Chloroflexus aurantiacus | |
| Homo sapiens (human) | |
| Glycine max (soybean) |
Taxonomic Hierarchy
GlycoGene Database (GGDB)
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| B4GALT5 | (not applicable) |
 G71142DF
G71142DF
|
Ceramide |
 G84224TW
G84224TW
|
Ceramide | |
| B4GALT6 | UDP-Gal |
 G71142DF
G71142DF
|
Cer |
 G84224TW
G84224TW
|
Cer | |
| GXYLT1 | UDP-Xyl |
 G71142DF
G71142DF
|
Ser |
 G01629YL
G01629YL
|
Ser | |
| GXYLT2 | UDP-Xyl |
 G71142DF
G71142DF
|
Ser |
 G01629YL
G01629YL
|
Ser |
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| B3GALT1 | UDP-Gal |
 G71142DF
G71142DF
|
Cer |
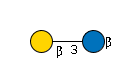 G31761VE
G31761VE
|
Cer | |
| B4GALT3 | UDP-Gal |
 G71142DF
G71142DF
|
[beta]1-Cer |
 G84224TW
G84224TW
|
[beta]1-Cer | |
| GXYLT1 | UDP-Xyl |
 G71142DF
G71142DF
|
Ser |
 G01629YL
G01629YL
|
Ser | |
| GXYLT2 | UDP-Xyl |
 G71142DF
G71142DF
|
Ser |
 G01629YL
G01629YL
|
Ser |
KEGG BRITE Database
- KEGG ID
- Enzyme
- Reactions
Pathway
| Pathway Name | Organism |
|---|---|
| E.coli O21 | Escherichia coli |
| E.coli O24 | Escherichia coli |
| E.coli O25 | Escherichia coli |
| E.coli O27 | Escherichia coli |
| E.coli O29 | Escherichia coli |
| E.coli O30 | Escherichia coli |
| E.coli O41 | Escherichia coli |
| E.coli O45 | Escherichia coli |
| E.coli O45rel | Escherichia coli |
| E.coli O46 | Escherichia coli |
Sequence Descriptors
- GlycoCT
-
RES 1b:b-dglc-HEX-1:5
- WURCS
- WURCS=2.0/1,1,0/[a2122h-1b_1-5]/1/
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 36124253 | Characterization and photocatalytic activity of TiO2 nanoparticles on cotton fabrics, for antibacterial masks | Alvarez-Amparán MA | 2022 Sep 15 |
|
| 35643637 | Cryo-EM structure and rRNA modification sites of a plant ribosome | Cottilli P | 2022 Sep 12 |
|
| 36088521 | Genome Resequencing and Transcriptome Analysis Reveal the Genetic Diversity of Wolfiporia cocos Germplasm and Genes Related to High Yield | Wang Q | 2022 Sep 10 |
|
| 35997626 | Enzyme kinetics by <scp>GH7</scp> cellobiohydrolases on chromogenic substrates is dictated by non‐productive binding: insights from crystal structures and <scp>MD</scp> simulation | Haataja T | 2022 Sep 06 |
|
| 36063177 | Reactive oxygen species in plants: an invincible fulcrum for biotic stress mitigation | Tyagi S | 2022 Sep 05 |
|
| 36069444 | Deciphering Cellodextrin and Glucose Uptake in Clostridium thermocellum | Yan F | 2022 Oct 26 |
|
| 36251112 | Synergistic Degradation of Maize Straw Lignin by Manganese Peroxidase from Irpex lacteus | Chen H | 2022 Oct 17 |
|
| 36241938 | β-glucosidase production by recombinant Pichia pastoris strain Y1433 under optimal feed profiles of fed-batch cultivation | Changming S | 2022 Oct 14 |
|
| 36197914 | Role of Half-of-Sites Reactivity and Inter-Subunit Communications in DAHP Synthase Catalysis and Regulation | Balachandran N | 2022 Oct 05 |
|
| 36618034 | Characterization and identification of inulin from Pachyrhizus erosus and evaluation of its antioxidant and in-vitro prebiotic efficacy | Bhanja A | 2022 Nov 20 |
|
 G80753ZP
G80753ZP
 G58161NS
G58161NS
 G24620AI
G24620AI
 G98256YA
G98256YA
 G72079JG
G72079JG
