Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE G71142DF
G71142DF
Summary
- GlyTouCan ID
-
G71142DF
- IUPAC Condensed
- Glc(b1-
- Motifs
- Glucosylceramide
External Links
- GlyGen
- ChEBI
- PubChem Compound
- PubChem Substance
- The Human Metabolome Database
3D Structures
- GlycoNAVI
- G71142DF
- GLYCAM
- DGlcpb1-OH
Organisms
| Organisms | Evidence |
|---|---|
| Mus musculus (house mouse) | |
| Clostridium | |
| Chloroflexus aurantiacus | |
| Homo sapiens (human) | |
| Glycine max (soybean) |
Taxonomic Hierarchy
GlycoGene Database (GGDB)
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| B4GALT5 | (not applicable) |
 G71142DF
G71142DF
|
Ceramide |
 G84224TW
G84224TW
|
Ceramide | |
| B4GALT6 | UDP-Gal |
 G71142DF
G71142DF
|
Cer |
 G84224TW
G84224TW
|
Cer | |
| GXYLT1 | UDP-Xyl |
 G71142DF
G71142DF
|
Ser |
 G01629YL
G01629YL
|
Ser | |
| GXYLT2 | UDP-Xyl |
 G71142DF
G71142DF
|
Ser |
 G01629YL
G01629YL
|
Ser |
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| B3GALT1 | UDP-Gal |
 G71142DF
G71142DF
|
Cer |
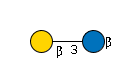 G31761VE
G31761VE
|
Cer | |
| B4GALT3 | UDP-Gal |
 G71142DF
G71142DF
|
[beta]1-Cer |
 G84224TW
G84224TW
|
[beta]1-Cer | |
| GXYLT1 | UDP-Xyl |
 G71142DF
G71142DF
|
Ser |
 G01629YL
G01629YL
|
Ser | |
| GXYLT2 | UDP-Xyl |
 G71142DF
G71142DF
|
Ser |
 G01629YL
G01629YL
|
Ser |
KEGG BRITE Database
- KEGG ID
- Enzyme
- Reactions
Pathway
| Pathway Name | Organism |
|---|---|
| E.coli O102 | Escherichia coli |
| E.coli O103 | Escherichia coli |
| E.coli O130 | Escherichia coli |
| E.coli O134 | Escherichia coli |
| E.coli O137 | Escherichia coli |
| E.coli O139 | Escherichia coli |
| E.coli O141 | Escherichia coli |
| E.coli O150 | Escherichia coli |
| E.coli O152 | Escherichia coli |
| E.coli O157 | Escherichia coli |
Sequence Descriptors
- GlycoCT
-
RES 1b:b-dglc-HEX-1:5
- WURCS
- WURCS=2.0/1,1,0/[a2122h-1b_1-5]/1/
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 34363539 | Colorimetric determination of phenolic compounds using peroxidase mimics based on biomolecule-free hybrid nanoflowers consisting of graphitic carbon nitride and copper | Dang TV | 2021 Aug 07 |
|
| 34347930 | The conserved C2 phospholipid‐binding domain in Delta contributes to robust Notch signalling | Martins T | 2021 Aug 04 |
|
| 34345982 | The phenolic acids from Oplopanax elatus Nakai stems and their potential photo-damage prevention activity | Han Y | 2021 Aug 03 |
|
| 35299916 | Effect of Senna plant on the mitochondrial activity of Hymenolepis diminuta | Ukil B | 2021 Aug 02 |
|
| 33941071 | D-Rhamnan and Pyruvate-Containing Teichuronic Acid from the Cell Wall of Rathayibacter sp. VKM Ac-2759 | Shashkov AS | 2021 Apr 09 |
|
| 33941069 | Heterologous Expression of Thermogutta terrifontis Endo-Xanthanase in Penicillium verruculosum, Isolation and Primary Characterization of the Enzyme | Denisenko YA | 2021 Apr 09 |
|
| 33820910 | Structure of the mannose phosphotransferase system (man-PTS) complexed with microcin E492, a pore-forming bacteriocin | Huang K | 2021 Apr 06 |
|
| 33791835 | Evaluation of the roles of hydrophobic residues in the N-terminal region of archaeal trehalase in its folding | Sakaguchi M | 2021 Apr 01 |
|
| 33422634 | A novel synergistic anticancer effect of fungal cholestanol glucoside and paclitaxel: Apoptosis induced by an intrinsic pathway through ROS generation in cervical cancer cell line (HeLa) | Subramaniam Y | 2021 Apr |
|
| 32945974 | Impact of non-proteinogenic amino acids in the discovery and development of peptide therapeutics | Ding Y | 2020 Sep 18 |
|
 G80753ZP
G80753ZP
 G58161NS
G58161NS
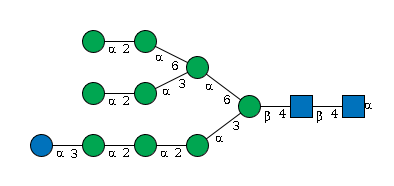 G24620AI
G24620AI
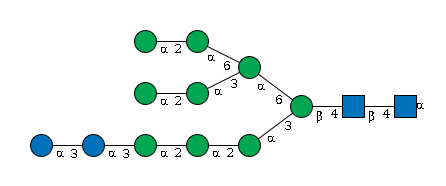 G98256YA
G98256YA
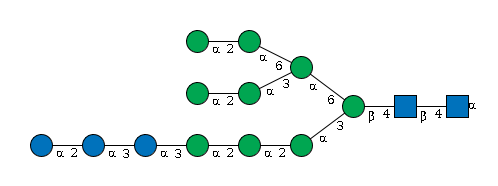 G72079JG
G72079JG
