Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE G71142DF
G71142DF
Summary
- GlyTouCan ID
-
G71142DF
- IUPAC Condensed
- Glc(b1-
- Motifs
- Glucosylceramide
External Links
- GlyGen
- ChEBI
- PubChem Compound
- PubChem Substance
- The Human Metabolome Database
3D Structures
- GlycoNAVI
- G71142DF
- GLYCAM
- DGlcpb1-OH
Organisms
| Organisms | Evidence |
|---|---|
| Mus musculus (house mouse) | |
| Clostridium | |
| Chloroflexus aurantiacus | |
| Homo sapiens (human) | |
| Glycine max (soybean) |
Taxonomic Hierarchy
GlycoGene Database (GGDB)
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| B4GALT5 | (not applicable) |
 G71142DF
G71142DF
|
Ceramide |
 G84224TW
G84224TW
|
Ceramide | |
| B4GALT6 | UDP-Gal |
 G71142DF
G71142DF
|
Cer |
 G84224TW
G84224TW
|
Cer | |
| GXYLT1 | UDP-Xyl |
 G71142DF
G71142DF
|
Ser |
 G01629YL
G01629YL
|
Ser | |
| GXYLT2 | UDP-Xyl |
 G71142DF
G71142DF
|
Ser |
 G01629YL
G01629YL
|
Ser |
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| B3GALT1 | UDP-Gal |
 G71142DF
G71142DF
|
Cer |
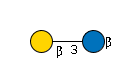 G31761VE
G31761VE
|
Cer | |
| B4GALT3 | UDP-Gal |
 G71142DF
G71142DF
|
[beta]1-Cer |
 G84224TW
G84224TW
|
[beta]1-Cer | |
| GXYLT1 | UDP-Xyl |
 G71142DF
G71142DF
|
Ser |
 G01629YL
G01629YL
|
Ser | |
| GXYLT2 | UDP-Xyl |
 G71142DF
G71142DF
|
Ser |
 G01629YL
G01629YL
|
Ser |
KEGG BRITE Database
- KEGG ID
- Enzyme
- Reactions
Pathway
| Pathway Name | Organism |
|---|---|
| E.coli O102 | Escherichia coli |
| E.coli O103 | Escherichia coli |
| E.coli O130 | Escherichia coli |
| E.coli O134 | Escherichia coli |
| E.coli O137 | Escherichia coli |
| E.coli O139 | Escherichia coli |
| E.coli O141 | Escherichia coli |
| E.coli O150 | Escherichia coli |
| E.coli O152 | Escherichia coli |
| E.coli O157 | Escherichia coli |
Sequence Descriptors
- GlycoCT
-
RES 1b:b-dglc-HEX-1:5
- WURCS
- WURCS=2.0/1,1,0/[a2122h-1b_1-5]/1/
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 32381508 | Structure–function analysis of silkworm sucrose hydrolase uncovers the mechanism of substrate specificity in GH13 subfamily 17 exo-α-glucosidases | Miyazaki T | 2020 Jun 26 |
|
| 32555346 | Structural and enzymatic characterisation of the Type III effector NopAA (=GunA) from Sinorhizobium fredii USDA257 reveals a Xyloglucan hydrolase activity | Dorival J | 2020 Jun 18 |
|
| 32514754 | Advances on the in vivo and in vitro glycosylations of flavonoids | Ji Y | 2020 Jun 08 |
|
| 32506169 | A comprehensive overview of substrate specificity of glycoside hydrolases and transporters in the small intestine : 'A gut feeling' | Elferink H | 2020 Jun 06 |
|
| 32666305 | Impact of Nutritional Stress on Honeybee Gut Microbiota, Immunity, and Nosema ceranae Infection | Castelli L | 2020 Jul 15 |
|
| 32666141 | Simultaneous voltammetric detection of glucose and lactate fluctuations in rat striatum evoked by electrical stimulation of the midbrain | Forderhase AG | 2020 Jul 14 |
|
| 32404367 | The bacterial metalloprotease NleD selectively cleaves mitogen-activated protein kinases that have high flexibility in their activation loop | Gur-Arie L | 2020 Jul 10 |
|
| 33505081 | Production of β-glucosidase from okara fermentation using Kluyveromyces marxianus | Su M | 2020 Jul 08 |
|
| 32559333 | Heptabladed β‐propeller lectins PLL2 and PHL from Photorhabdus spp. recognize O‐methylated sugars and influence the host immune system | Fujdiarová E | 2020 Jul 07 |
|
| 32647570 | Changes in the gut microbiota mediate the differential regulatory effects of two glucose oxidases produced by Aspergillus niger and Penicillium amagasakiense on the meat quality and growth performance of broilers | Wu S | 2020 Jul 06 |
|
 G80753ZP
G80753ZP
 G58161NS
G58161NS
 G24620AI
G24620AI
 G98256YA
G98256YA
 G72079JG
G72079JG
