Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE G71142DF
G71142DF
Summary
- GlyTouCan ID
-
G71142DF
- IUPAC Condensed
- Glc(b1-
- Motifs
- Glucosylceramide
External Links
- GlyGen
- ChEBI
- PubChem Compound
- PubChem Substance
- The Human Metabolome Database
3D Structures
- GlycoNAVI
- G71142DF
- GLYCAM
- DGlcpb1-OH
Organisms
| Organisms | Evidence |
|---|---|
| Mus musculus (house mouse) | |
| Clostridium | |
| Chloroflexus aurantiacus | |
| Homo sapiens (human) | |
| Glycine max (soybean) |
Taxonomic Hierarchy
GlycoGene Database (GGDB)
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| B4GALT5 | (not applicable) |
 G71142DF
G71142DF
|
Ceramide |
 G84224TW
G84224TW
|
Ceramide | |
| B4GALT6 | UDP-Gal |
 G71142DF
G71142DF
|
Cer |
 G84224TW
G84224TW
|
Cer | |
| GXYLT1 | UDP-Xyl |
 G71142DF
G71142DF
|
Ser |
 G01629YL
G01629YL
|
Ser | |
| GXYLT2 | UDP-Xyl |
 G71142DF
G71142DF
|
Ser |
 G01629YL
G01629YL
|
Ser |
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| B4GALT5 | (not applicable) |
 G71142DF
G71142DF
|
Ceramide |
 G84224TW
G84224TW
|
Ceramide | |
| B3GALT2 | UDP-Gal |
 G71142DF
G71142DF
|
Cer |
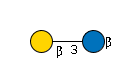 G31761VE
G31761VE
|
Cer | |
| B4GALT1 | UDP-Gal |
 G71142DF
G71142DF
|
R |
 G84224TW
G84224TW
|
R | |
| B4GALT6 | UDP-Gal |
 G71142DF
G71142DF
|
 G84224TW
G84224TW
|
|||
| B4GALT6 | UDP-Gal |
 G71142DF
G71142DF
|
Cer |
 G84224TW
G84224TW
|
Cer |
KEGG BRITE Database
- KEGG ID
- Enzyme
- Reactions
Pathway
| Pathway Name | Organism |
|---|---|
| E.coli O21 | Escherichia coli |
| E.coli O24 | Escherichia coli |
| E.coli O25 | Escherichia coli |
| E.coli O27 | Escherichia coli |
| E.coli O29 | Escherichia coli |
| E.coli O30 | Escherichia coli |
| E.coli O41 | Escherichia coli |
| E.coli O45 | Escherichia coli |
| E.coli O45rel | Escherichia coli |
| E.coli O46 | Escherichia coli |
Sequence Descriptors
- GlycoCT
-
RES 1b:b-dglc-HEX-1:5
- WURCS
- WURCS=2.0/1,1,0/[a2122h-1b_1-5]/1/
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 33231837 | Amyloid β aggregation inhibitory activity of triterpene saponins from the cactus Stenocereus pruinosus | Fujihara K | 2020 Nov 24 |
|
| 33208356 | Na + -dependent gate dynamics and electrostatic attraction ensure substrate coupling in glutamate transporters | Alleva C | 2020 Nov 20 |
|
| 33208922 | Dietary fibre in gastrointestinal health and disease | Gill SK | 2020 Nov 18 |
|
| 33188507 | Repurposing Inflatable Packaging Pillows as Bioreactors: a Convenient Synthesis of Glucosone by Whole-Cell Catalysis Under Oxygen | Mozuch MD | 2020 Nov 13 |
|
| 33155685 | A non‐helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition | Focht D | 2020 Nov 06 |
|
| 33146790 | Status of the application of exogenous enzyme technology for the development of natural plant resources | Yuan B | 2020 Nov 04 |
|
| 33141351 | Genome sequence of an uncharted halophilic bacterium Robertkochia marina with deciphering its phosphate-solubilizing ability | Lam MQ | 2020 Nov 03 |
|
| 32451508 | Structural insights into β-1,3-glucan cleavage by a glycoside hydrolase family | Santos CR | 2020 May 25 |
|
| 32449090 | Characterization of FsXEG12A from the cellulose-degrading ectosymbiotic fungus Fusarium spp. strain EI cultured by the ambrosia beetle | Sakai K | 2020 May 24 |
|
| 32424254 | Aerobic radical polymerization mediated by microbial metabolism | Fan G | 2020 May 18 |
|
 G80753ZP
G80753ZP
 G58161NS
G58161NS
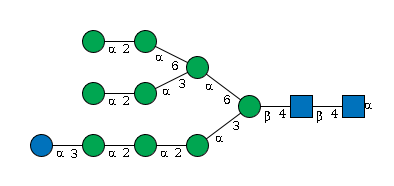 G24620AI
G24620AI
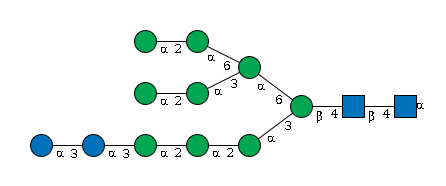 G98256YA
G98256YA
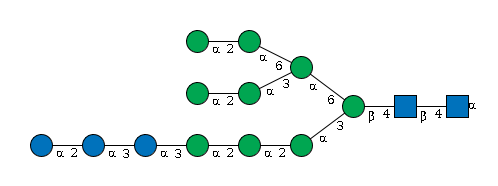 G72079JG
G72079JG
