Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
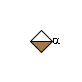
MIRAGE G79824WM
G79824WM
Summary
- GlyTouCan ID
-
G79824WM
- IUPAC Condensed
- IdoA(a1-
Pathway
| Pathway Name | Organism |
|---|---|
| E.coli O112ab | Escherichia coli |
Sequence Descriptors
- GlycoCT
-
RES 1b:a-lido-HEX-1:5|6:a
- WURCS
- WURCS=2.0/1,1,0/[a2121A-1a_1-5]/1/
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 38430291 | Characterization and Biological Activities of the Ulvan Polysaccharide-Rich Fraction Obtained from Ulva rigida and Ulva pseudorotundata and Their Potential for Pharmaceutical Application | Fávero Massocato T | 2024 Mar 02 |
|
| 37843732 | Protein-modified nanomaterials: emerging trends in skin wound healing | Sharda D | 2023 Oct 16 |
|
| 38765689 | Biochemical characterisation of a PL24 ulvan lyase from seaweed-associated Vibrio sp. FNV38 | Rodrigues VJ | 2023 Dec 07 |
|
| 34626405 | Computerized Molecular ModelingMolecular modeling for Discovering Promising Glycosaminoglycan OligosaccharidesOligosaccharides that Modulate Protein\nProteins\n Function | Sankaranarayanan NV | 2022 |
|
| 34194082 | Sulfated polysaccharides and its commercial applications in food industries—A review | Muthukumar J | 2020 Oct 15 |
|
| 32274709 | An Overview of the Structure, Mechanism and Specificity of Human Heparanase | Wu L | 2020 |
|
| 32274740 | The Good and Bad Sides of Heparanase-1 and Heparanase-2 | Pinhal MAS | 2020 |
|
| 31061498 | Glycan sulfation patterns define autophagy flux at axon tip via PTPRσ-cortactin axis | Sakamoto K | 2019 May 06 |
|
| 30061628 | Disease and subtype specific signatures enable precise diagnosis of the mucopolysaccharidoses | Saville JT | 2019 Mar |
|
| 29980923 | Demystifying the pH dependent conformational changes of human heparanase pertaining to structure–function relationships: an in silico approach | Nagarajan H | 2018 Jul 06 |
|