Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE G00056MO
G00056MO
3D Structures
- GlycoNAVI
- G00056MO
- GlycoShape
- G00056MO
- GLYCAM
- DGalpb1-3DGlcpNAcb1-OH
Organisms
| Organisms | Evidence |
|---|---|
| Helicobacter pylori | |
| Homo sapiens (human) | |
| Escherichia coli |
Taxonomic Hierarchy
GlycoGene Database (GGDB)
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| FUT2 | GDP-Fuc |
 G00056MO
G00056MO
|
Free |
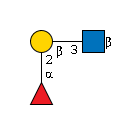 G40696TO
G40696TO
|
Free | |
| FUT5 | GDP-Fuc |
 G00056MO
G00056MO
|
R |
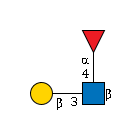 G39023AU
G39023AU
|
R | |
| FUT1 | GDP-Fuc |
 G00056MO
G00056MO
|
Free |
 G40696TO
G40696TO
|
Free | |
| FUT3 | GDP-Fuc |
 G00056MO
G00056MO
|
R |
 G39023AU
G39023AU
|
R | |
| B3GALT1 | UDP-Gal |
 G49108TO
G49108TO
|
Lemieux |
 G00056MO
G00056MO
|
Lemieux |
Pathway
| Pathway Name | Organism |
|---|---|
| E.coli O174ac | Escherichia coli |
| E.coli O64 | Escherichia coli |
Sequence Descriptors
- GlycoCT
-
RES 1b:b-dglc-HEX-1:5 2s:n-acetyl 3b:b-dgal-HEX-1:5 LIN 1:1d(2+1)2n 2:1o(3+1)3d
- WURCS
- WURCS=2.0/2,2,1/[a2122h-1b_1-5_2*NCC/3=O][a2112h-1b_1-5]/1-2/a3-b1
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 32620774 | Butyrate producing colonic Clostridiales metabolise human milk oligosaccharides and cross feed on mucin via conserved pathways | Pichler MJ | 2020 Jul 03 |
|
| 31118252 | GII.13/21 noroviruses recognize glycans with a terminal beta-galactose via an unconventional glycan binding site | Cong X | 2019 May 22 |
|
| 31226167 | Unraveling the role of the secretor antigen in human rotavirus attachment to histo-blood group antigens | Gozalbo-Rovira R | 2019 Jun 12 |
|
| 29848588 | Structural and Molecular Characterization of the Hemagglutinin from the Fifth-Epidemic-Wave A(H7N9) Influenza Viruses | Yang H | 2018 Aug 15 |
|
| 28392148 | Molecular Insight into Evolution of Symbiosis between Breast-Fed Infants and a Member of the Human Gut Microbiome Bifidobacterium longum | Yamada C | 2017 Apr 20 |
|
| 28000747 | Structure-based rationale for differential recognition of lacto- and neolacto- series glycosphingolipids by the N-terminal domain of human galectin-8 | Bohari MH | 2016 Dec 21 |
|
| 25945972 | Structural Basis Underlying the Binding Preference of Human Galectins-1, -3 and -7 for Galβ1-3/4GlcNAc | Hsieh T | 2015 May 06 |
|
| 26077389 | Structural characterization of human galectin‐4 C‐terminal domain: elucidating the molecular basis for recognition of glycosphingolipids, sulfated saccharides and blood group antigens | Bum-Erdene K | 2015 Jul 14 |
|
| 26105055 | Structural Hot Spots Determine Functional Diversity of the Candida glabrata Epithelial Adhesin Family | Diderrich R | 2015 Aug 07 |
|
| 25673707 | Structure and Receptor Binding Preferences of Recombinant Hemagglutinins from Avian and Human H6 and H10 Influenza A Virus Subtypes | Yang H | 2015 Apr 15 |
|
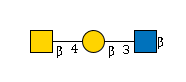 G52935RP
G52935RP
