Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE G16579HJ
G16579HJ
Summary
- GlyTouCan ID
-
G16579HJ
- IUPAC Condensed
- Xyl(b1-
External Links
- GlyGen
- ChEBI
- PubChem Compound
- PubChem Substance
- CarbBank
- 2180 2181 3939 3984 23404 26445 26446 26447 26448 26993 27125 27150 33278 33279 34065 36092 36093 37941 38733 38766 38849 38932 39011 39012 39013 39014 39053 39069 39866 39867 39907 40096 40097 40131 41171 41409 42546 42547 42548 43699 43700 43901 43907 43942 44766 44868 44913 46100 46101 46102 46441 48293 48772 48800 48801 48802 48803 49287 49568 49733 49748 49749 49750 49751 49752 49753 49754 49755 49756 49760 49815 49819 50045 50081 50569 50570
3D Structures
- GLYCAM
- DXylpb1-OH
Organisms
| Organisms | Evidence |
|---|---|
| Gymnocladus dioicus (Kentucky coffee tree) | |
| Solidago virgaurea | |
| Astragalus penduliflorus | |
| Gymnocladus chinensis | |
| Asteroidea (starfishes) |
Taxonomic Hierarchy
GlycoGene Database (GGDB)
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| B4GALT7 | (not applicable) |
 G16579HJ
G16579HJ
|
R |
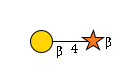 G78098IA
G78098IA
|
R | |
| B4GALT7 | UDP-Gal |
 G16579HJ
G16579HJ
|
4-D-MethylUmb-[beta] |
 G78098IA
G78098IA
|
4-D-MethylUmb-[beta] | |
| B4GALT7 | UDP-Gal |
 G16579HJ
G16579HJ
|
O-Nitrophenyl-[beta] |
 G78098IA
G78098IA
|
O-Nitrophenyl-[beta] | |
| B4GALT7 | UDP-Gal |
 G16579HJ
G16579HJ
|
proteoglycan core |
 G78098IA
G78098IA
|
proteoglycan core | |
| B4GAT1 | UDP-GlcA |
 G16579HJ
G16579HJ
|
R |
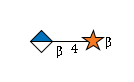 G42293RF
G42293RF
|
R |
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Reference |
|---|---|---|---|---|
| B3GALT6 | UDP-Gal |
 G16579HJ
G16579HJ
|
Benzyl, naphthol | |
| B4GALT7 | UDP-Gal |
 G16579HJ
G16579HJ
|
Methyl | |
| B4GALT7 | UDP-Gal |
 G16579HJ
G16579HJ
|
p-Nitrophenyl-[alpha] | |
| B4GALT7 | UDP-Gal |
 G16579HJ
G16579HJ
|
p-Nitrophenyl-3 | |
| B4GALT4 | UDP-Gal |
 G16579HJ
G16579HJ
|
[beta]-1-4-methyl-umbelliferyl |
KEGG BRITE Database
Sequence Descriptors
- GlycoCT
-
RES 1b:b-dxyl-PEN-1:5
- WURCS
- WURCS=2.0/1,1,0/[a212h-1b_1-5]/1/
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 37715802 | Investigating the effect of substrate binding on the catalytic activity of xylanase | Ma L | 2023 Sep 16 |
|
| 37932829 | Impact of xylose epimerase on sugar assimilation and sensing in recombinant Saccharomyces cerevisiae carrying different xylose-utilization pathways | Persson VC | 2023 Nov 06 |
|
| 37921982 | A comprehensive review on genetic modification of plant cell wall for improved saccharification efficiency | Mishra A | 2023 Nov 03 |
|
| 37144255 | Crystal structure of reducing‐end xylose‐releasing exoxylanase in subfamily 7 of glycoside hydrolase family 30 | Nakamichi Y | 2023 May 05 |
|
| 38765689 | Biochemical characterisation of a PL24 ulvan lyase from seaweed-associated Vibrio sp. FNV38 | Rodrigues VJ | 2023 Dec 07 |
|
| 37561381 | Revealing the Potential Impacts of Nutraceuticals Formulated with Freeze-Dried Jabuticaba Peel and Limosilactobacillus fermentum Strains Candidates for Probiotic Use on Human Intestinal Microbiota | da Silva JYP | 2023 Aug 10 |
|
| 37093530 | Optimization and Physicochemical Characterization of Polysaccharide Purified from Sonneratia caseolaris Mangrove Leaves: a Potential Antioxidant and Antibiofilm Agent | Jha N | 2023 Apr 24 |
|
| 38647872 | An accessory enzymatic system of cellulase for simultaneous saccharification and co-fermentation | Liu H | 2022 Sep 19 |
|
| 35398092 | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases | Cross AR | 2022 May |
|
| 35120929 | Comparison of glycoside hydrolase family 3 β-xylosidases from basidiomycetes and ascomycetes reveals evolutionarily distinct xylan degradation systems | Kojima K | 2022 Mar |
|
 G75592IR
G75592IR
 G03144EF
G03144EF
