Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE G16579HJ
G16579HJ
Summary
- GlyTouCan ID
-
G16579HJ
- IUPAC Condensed
- Xyl(b1-
External Links
- GlyGen
- ChEBI
- PubChem Compound
- PubChem Substance
- CarbBank
- 2180 2181 3939 3984 23404 26445 26446 26447 26448 26993 27125 27150 33278 33279 34065 36092 36093 37941 38733 38766 38849 38932 39011 39012 39013 39014 39053 39069 39866 39867 39907 40096 40097 40131 41171 41409 42546 42547 42548 43699 43700 43901 43907 43942 44766 44868 44913 46100 46101 46102 46441 48293 48772 48800 48801 48802 48803 49287 49568 49733 49748 49749 49750 49751 49752 49753 49754 49755 49756 49760 49815 49819 50045 50081 50569 50570
3D Structures
- GLYCAM
- DXylpb1-OH
Organisms
| Organisms | Evidence |
|---|---|
| Gymnocladus dioicus (Kentucky coffee tree) | |
| Solidago virgaurea | |
| Astragalus penduliflorus | |
| Gymnocladus chinensis | |
| Asteroidea (starfishes) |
Taxonomic Hierarchy
GlycoGene Database (GGDB)
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| B4GALT7 | (not applicable) |
 G16579HJ
G16579HJ
|
R |
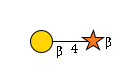 G78098IA
G78098IA
|
R | |
| B4GALT7 | UDP-Gal |
 G16579HJ
G16579HJ
|
4-D-MethylUmb-[beta] |
 G78098IA
G78098IA
|
4-D-MethylUmb-[beta] | |
| B4GALT7 | UDP-Gal |
 G16579HJ
G16579HJ
|
O-Nitrophenyl-[beta] |
 G78098IA
G78098IA
|
O-Nitrophenyl-[beta] | |
| B4GALT7 | UDP-Gal |
 G16579HJ
G16579HJ
|
proteoglycan core |
 G78098IA
G78098IA
|
proteoglycan core | |
| B4GAT1 | UDP-GlcA |
 G16579HJ
G16579HJ
|
R |
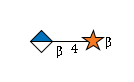 G42293RF
G42293RF
|
R |
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Reference |
|---|---|---|---|---|
| B3GALT6 | UDP-Gal |
 G16579HJ
G16579HJ
|
Benzyl, naphthol | |
| B4GALT7 | UDP-Gal |
 G16579HJ
G16579HJ
|
Methyl | |
| B4GALT7 | UDP-Gal |
 G16579HJ
G16579HJ
|
p-Nitrophenyl-[alpha] | |
| B4GALT7 | UDP-Gal |
 G16579HJ
G16579HJ
|
p-Nitrophenyl-3 | |
| B4GALT4 | UDP-Gal |
 G16579HJ
G16579HJ
|
[beta]-1-4-methyl-umbelliferyl |
KEGG BRITE Database
Sequence Descriptors
- GlycoCT
-
RES 1b:b-dxyl-PEN-1:5
- WURCS
- WURCS=2.0/1,1,0/[a212h-1b_1-5]/1/
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 35120456 | Profiling the cell walls of seagrasses from A (Amphibolis) to Z (Zostera) | Pfeifer L | 2022 Feb 04 |
|
| 38647596 | Production of cellulosic ethanol and value-added products from corn fiber | Guo Y | 2022 Aug 13 |
|
| 34626391 | Capillary Electrophoretic Analysis of Isolated Sulfated Polysaccharides to Characterize Pharmaceutical Products | Shriver Z | 2022 |
|
| 34499201 | Herbaspirillum seropedicae expresses non-phosphorylative pathways for d-xylose catabolism | Malán AK | 2021 Sep 09 |
|
| 34790504 | Nanomaterial conjugated lignocellulosic waste: cost-effective production of sustainable bioenergy using enzymes | Kaur P | 2021 Oct 30 |
|
| 34838122 | Valorization of Chinese hickory shell as novel sources for the efficient production of xylooligosaccharides | Wang Z | 2021 Nov 27 |
|
| 34761460 | Three‐dimensional structure of xylonolactonase from Caulobacter crescentus: A mononuclear iron enzyme of the 6‐bladed β‐propeller hydrolase family | Pääkkönen J | 2021 Nov 20 |
|
| 33446650 | Two distinct catalytic pathways for GH43 xylanolytic enzymes unveiled by X-ray and QM/MM simulations | Morais MAB | 2021 Jan 14 |
|
| 36304480 | Cell size: a key determinant of meristematic potential in plant protoplasts | Pujari I | 2021 Jan 01 |
|
| 33242495 | Crystal structure of Dictyoglomus thermophilum β-d-xylosidase DtXyl unravels the structural determinants for efficient notoginsenoside R1 hydrolysis | Bretagne D | 2021 Feb |
|
 G75592IR
G75592IR
 G03144EF
G03144EF
