Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE G49108TO
G49108TO
Summary
- GlyTouCan ID
-
G49108TO
- IUPAC Condensed
- GlcNAc(b1-
3D Structures
- GLYCAM
- DGlcpNAcb1-OH
Organisms
| Organisms | Evidence |
|---|---|
| Mus musculus (house mouse) | |
| Rattus norvegicus (Norway rat) | |
| Homo sapiens (human) | |
| Drosophila melanogaster (fruit fly) | |
| Streptococcus pneumoniae |
Taxonomic Hierarchy
GlycoGene Database (GGDB)
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| B4GALT5 | (not applicable) |
 G49108TO
G49108TO
|
[beta]-S-pNP |
 G00055MO
G00055MO
|
[beta]-S-pNP | |
| B3GALT2 | UDP-Gal |
 G49108TO
G49108TO
|
Lemieux |
 G00056MO
G00056MO
|
Lemieux | |
| B4GALT2 | UDP-Gal |
 G49108TO
G49108TO
|
Benzyl-[beta] |
 G00055MO
G00055MO
|
Benzyl-[beta] | |
| B4GALT1 | UDP-Gal |
 G49108TO
G49108TO
|
R |
 G00055MO
G00055MO
|
R | |
| B3GALT5 | UDP-Gal |
 G49108TO
G49108TO
|
R |
 G00056MO
G00056MO
|
R |
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| B3GALT5 | UDP-Gal |
 G49108TO
G49108TO
|
R |
 G00056MO
G00056MO
|
R | |
| B3GALT1 | UDP-Gal |
 G49108TO
G49108TO
|
Lemieux |
 G00056MO
G00056MO
|
Lemieux | |
| B4GALT4 | UDP-Gal |
 G49108TO
G49108TO
|
[beta]-1-thio-p-Nitrophenyl |
 G00055MO
G00055MO
|
[beta]-1-thio-p-Nitrophenyl | |
| B4GALNT3 | UDP-GalNAc |
 G49108TO
G49108TO
|
R |
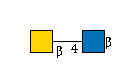 G00057MO
G00057MO
|
R | |
| B3GALNT2 | UDP-GalNAc |
 G49108TO
G49108TO
|
[beta]-Bz |
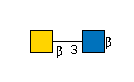 G36387CV
G36387CV
|
[beta]-Bz |
Core Protein
GlycoEpitope
- Epitope ID
- EP0004
Pathway
| Pathway Name | Organism |
|---|---|
| Antimicrobial peptides | Rattus norvegicus |
| E.coli O10 | Escherichia coli |
| E.coli O103 | Escherichia coli |
| E.coli O105 | Escherichia coli |
| E.coli O109 | Escherichia coli |
| E.coli O110 | Escherichia coli |
| E.coli O111 | Escherichia coli |
| E.coli O112ab | Escherichia coli |
| E.coli O113 | Escherichia coli |
| E.coli O115 | Escherichia coli |
Sequence Descriptors
- GlycoCT
-
RES 1b:b-dglc-HEX-1:5 2s:n-acetyl LIN 1:1d(2+1)2n
- WURCS
- WURCS=2.0/1,1,0/[a2122h-1b_1-5_2*NCC/3=O]/1/
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 35256819 | An anti-PD-1–GITR-L bispecific agonist induces GITR clustering-mediated T cell activation for cancer immunotherapy | Chan S | 2022 Mar 07 |
|
| 35253643 | How clustered protocadherin binding specificity is tuned for neuronal self-/nonself-recognition | Goodman KM | 2022 Mar 07 |
|
| 35253970 | Biparatopic sybodies neutralize SARS‐CoV‐2 variants of concern and mitigate drug resistance | Walter JD | 2022 Mar 07 |
|
| 35246509 | Evolution of the SARS-CoV-2 spike protein in the human host | Wrobel AG | 2022 Mar 04 |
|
| 35133176 | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody | Yin W | 2022 Mar 04 |
|
| 35120603 | Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron | Cui Z | 2022 Mar 03 |
|
| 35241675 | Cryo-EM structure of a SARS-CoV-2 omicron spike protein ectodomain | Ye G | 2022 Mar 03 |
|
| 35236939 | Structural basis of the activation of metabotropic glutamate receptor 3 | Fang W | 2022 Mar 02 |
|
| 35172173 | Cryo-EM structure of the SARS-CoV-2 Omicron spike | Cerutti G | 2022 Mar 01 |
|
| 35230385 | Epitope convergence of broadly HIV-1 neutralizing IgA and IgG antibody lineages in a viremic controller | Lorin V | 2022 Mar 01 |
|
PubAnnotation
| Title | Authors | Source | Publication Date | PubMed ID |
|---|---|---|---|---|
| Dynamic glycosylation of the transcription factor CREB: a potential role in gene regulation. |
|
J Am Chem Soc | 2003 Jun 4 | 12769553 |
| Effect of okadaic acid on O-linked N-acetylglucosamine levels in a neuroblastoma cell line. |
|
Biochim Biophys Acta | 1999 Oct 18 | 10572927 |
| Regulation of a cytosolic and nuclear O-GlcNAc transferase. Role of the tetratricopeptide repeats. |
|
J Biol Chem | 1999 Nov 5 | 10542233 |
| An N-terminal region of Sp1 targets its proteasome-dependent degradation in vitro. |
|
J Biol Chem | 1999 May 21 | 10329728 |
| Vertebrate lens alpha-crystallins are modified by O-linked N-acetylglucosamine. |
|
J Biol Chem | 1992 Jan 5 | 1730617 |
 G14843DJ
G14843DJ
 G68544GH
G68544GH
