Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE G49108TO
G49108TO
Summary
- GlyTouCan ID
-
G49108TO
- IUPAC Condensed
- GlcNAc(b1-
3D Structures
- GlycoNAVI
- G49108TO
- GLYCAM
- DGlcpNAcb1-OH
Organisms
| Organisms | Evidence |
|---|---|
| Mus musculus (house mouse) | |
| Rattus norvegicus (Norway rat) | |
| Homo sapiens (human) | |
| Drosophila melanogaster (fruit fly) | |
| Streptococcus pneumoniae |
Taxonomic Hierarchy
GlycoGene Database (GGDB)
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| B4GALT5 | (not applicable) |
 G49108TO
G49108TO
|
[beta]-S-pNP |
 G00055MO
G00055MO
|
[beta]-S-pNP | |
| B3GALT2 | UDP-Gal |
 G49108TO
G49108TO
|
Lemieux |
 G00056MO
G00056MO
|
Lemieux | |
| B4GALT2 | UDP-Gal |
 G49108TO
G49108TO
|
Benzyl-[beta] |
 G00055MO
G00055MO
|
Benzyl-[beta] | |
| B4GALT1 | UDP-Gal |
 G49108TO
G49108TO
|
R |
 G00055MO
G00055MO
|
R | |
| B3GALT5 | UDP-Gal |
 G49108TO
G49108TO
|
R |
 G00056MO
G00056MO
|
R |
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| B3GALT5 | UDP-Gal |
 G49108TO
G49108TO
|
R |
 G00056MO
G00056MO
|
R | |
| B3GALT1 | UDP-Gal |
 G49108TO
G49108TO
|
Lemieux |
 G00056MO
G00056MO
|
Lemieux | |
| B4GALT4 | UDP-Gal |
 G49108TO
G49108TO
|
[beta]-1-thio-p-Nitrophenyl |
 G00055MO
G00055MO
|
[beta]-1-thio-p-Nitrophenyl | |
| B4GALNT3 | UDP-GalNAc |
 G49108TO
G49108TO
|
R |
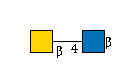 G00057MO
G00057MO
|
R | |
| B3GALNT2 | UDP-GalNAc |
 G49108TO
G49108TO
|
[beta]-Bz |
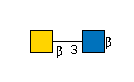 G36387CV
G36387CV
|
[beta]-Bz |
Core Protein
GlycoEpitope
- Epitope ID
- EP0004
Pathway
| Pathway Name | Organism |
|---|---|
| E.coli O28ab | Escherichia coli |
| E.coli O28ac | Escherichia coli |
| E.coli O29 | Escherichia coli |
| E.coli O3 | Escherichia coli |
| E.coli O30 | Escherichia coli |
| E.coli O32 | Escherichia coli |
| E.coli O35 | Escherichia coli |
| E.coli O36 | Escherichia coli |
| E.coli O37 | Escherichia coli |
| E.coli O38 | Escherichia coli |
Sequence Descriptors
- GlycoCT
-
RES 1b:b-dglc-HEX-1:5 2s:n-acetyl LIN 1:1d(2+1)2n
- WURCS
- WURCS=2.0/1,1,0/[a2122h-1b_1-5_2*NCC/3=O]/1/
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 3191922 | Investigation of protein structure by means of 19F-NMR. A study of hen egg-white lysozyme | Adriaensens P | 1988 Nov 01 |
|
| 3150939 | Zonal turnover of cell poles of Bacillus subtilis | Kirchner G | 1988 Nov |
|
| 3171165 | Visualization of exocellular lectins in the entomopathogenic fungus Conidiobolus obscurus. | Latgé JP | 1988 Nov |
|
| 3228136 | Aspartylglucosaminuria in a Puerto Rican family: Additional features of a panethnic disorder | Chitayat D | 1988 Nov |
|
| 3222257 | Lectin histochemistry of normal lung and pulmonary adenocarcinoma | Kawai T | 1988 Nov |
|
| 3143804 | Membrane Binding and Release of Bacillus subtilis DNA as a Function of the Cell Cycle | Sandler N | 1988 May 01 |
|
| 3368475 | Differential Mechanisms of Feeding Modulation Induced by Amino Sugars in Rats | Okabe Y | 1988 May 01 |
|
| 3349087 | Enzymatic synthesis of p-nitrophenyl N,N',N'',N'',N''''-pentaacetyl-beta-chitopentaoside in water-methanol system; significance as a substrate for lysozyme assay | Usui T | 1988 Mar 23 |
|
| 3370843 | Analysis of cell surface glycoconjugates in fibroblasts from patients with cystic fibrosis | Sarras MP | 1988 Mar 15 |
|
| 3126181 | Kluyveromyces bulgaricus yeast lectins. Isolation of N-acetylglucosamine and galactose-specific lectins: their relation with flocculation. | Al-Mahmood S | 1988 Mar 15 |
|
PubAnnotation
| Title | Authors | Source | Publication Date | PubMed ID |
|---|---|---|---|---|
| Phosphorylation activates master growth regulator DELLA by promoting histone H2A binding at chromatin in Arabidopsis. |
|
Nat Commun | 2024 Sep 3 | 39227587 |
| A protein O-GlcNAc glycosyltransferase regulates the antioxidative response in Yersinia pestis. |
|
Nat Commun | 2024 Aug 16 | 39152136 |
| Polη O-GlcNAcylation governs genome integrity during translesion DNA synthesis. |
|
Nat Commun | 2017 Dec 5 | 29208956 |
| Epithelial Mesenchymal Transition Induces Aberrant Glycosylation through Hexosamine Biosynthetic Pathway Activation. |
|
J Biol Chem | 2016 Jun 17 | 27129262 |
| The making of a sweet modification: structure and function of O-GlcNAc transferase. |
|
J Biol Chem | 2014 Dec 12 | 25336649 |
 G14843DJ
G14843DJ
 G68544GH
G68544GH
