Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE G49108TO
G49108TO
Summary
- GlyTouCan ID
-
G49108TO
- IUPAC Condensed
- GlcNAc(b1-
3D Structures
- GLYCAM
- DGlcpNAcb1-OH
Organisms
| Organisms | Evidence |
|---|---|
| Mus musculus (house mouse) | |
| Rattus norvegicus (Norway rat) | |
| Homo sapiens (human) | |
| Drosophila melanogaster (fruit fly) | |
| Streptococcus pneumoniae |
Taxonomic Hierarchy
GlycoGene Database (GGDB)
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| B4GALT5 | (not applicable) |
 G49108TO
G49108TO
|
[beta]-S-pNP |
 G00055MO
G00055MO
|
[beta]-S-pNP | |
| B3GALT2 | UDP-Gal |
 G49108TO
G49108TO
|
Lemieux |
 G00056MO
G00056MO
|
Lemieux | |
| B4GALT2 | UDP-Gal |
 G49108TO
G49108TO
|
Benzyl-[beta] |
 G00055MO
G00055MO
|
Benzyl-[beta] | |
| B4GALT1 | UDP-Gal |
 G49108TO
G49108TO
|
R |
 G00055MO
G00055MO
|
R | |
| B3GALT5 | UDP-Gal |
 G49108TO
G49108TO
|
R |
 G00056MO
G00056MO
|
R |
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| B3GALT5 | UDP-Gal |
 G49108TO
G49108TO
|
R |
 G00056MO
G00056MO
|
R | |
| B3GALT1 | UDP-Gal |
 G49108TO
G49108TO
|
Lemieux |
 G00056MO
G00056MO
|
Lemieux | |
| B4GALT4 | UDP-Gal |
 G49108TO
G49108TO
|
[beta]-1-thio-p-Nitrophenyl |
 G00055MO
G00055MO
|
[beta]-1-thio-p-Nitrophenyl | |
| B4GALNT3 | UDP-GalNAc |
 G49108TO
G49108TO
|
R |
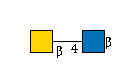 G00057MO
G00057MO
|
R | |
| B3GALNT2 | UDP-GalNAc |
 G49108TO
G49108TO
|
[beta]-Bz |
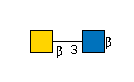 G36387CV
G36387CV
|
[beta]-Bz |
Core Protein
| UniProt ID | Protein Name | Reference | Source |
|---|---|---|---|
| A0A8I6A122 | TATA box-binding protein-like 1 | ||
| A0A8I6AMY7 | EMSY transcriptional repressor, BRCA2 interacting | ||
| A0AV96 | RNA-binding protein 47 | ||
| A0AVK6 | Transcription factor E2F8 | ||
| A0AVT1 | Ubiquitin-like modifier-activating enzyme 6 | ||
| A0FGR8 | Extended synaptotagmin-2 | ||
| A0MZ66 | Shootin-1 | ||
| A0PJY2 | Fez family zinc finger protein 1 | ||
| A1A6K6 | Nuclear speckle RNA-binding protein A | ||
| A1L0T0 | 2-hydroxyacyl-CoA lyase 2 |
GlycoEpitope
- Epitope ID
- EP0004
Pathway
| Pathway Name | Organism |
|---|---|
| Antimicrobial peptides | Xenopus tropicalis |
| Antimicrobial peptides | Rattus norvegicus |
| Antimicrobial peptides | Danio rerio |
| Antimicrobial peptides | Bos taurus |
| Antimicrobial peptides | Canis familiaris |
| Antimicrobial peptides | Sus scrofa |
| Antimicrobial peptides | Dictyostelium discoideum |
| Antimicrobial peptides | Mus musculus |
| Antimicrobial peptides | Drosophila melanogaster |
| Antimicrobial peptides | Homo sapiens |
Sequence Descriptors
- GlycoCT
-
RES 1b:b-dglc-HEX-1:5 2s:n-acetyl LIN 1:1d(2+1)2n
- WURCS
- WURCS=2.0/1,1,0/[a2122h-1b_1-5_2*NCC/3=O]/1/
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 35384608 | Efficient production of GlcNAc in an aqueous-organic system with a Chitinolyticbacter meiyuanensis SYBC-H1 mutant | Hao ZK | 2022 Apr 06 |
|
| 35230146 | Vaccination with SARS-CoV-2 spike protein lacking glycan shields elicits enhanced protective responses in animal models | Huang HY | 2022 Apr 06 |
|
| 35383176 | Structure-based design of stabilized recombinant influenza neuraminidase tetramers | Ellis D | 2022 Apr 05 |
|
| 35380894 | Cryoelectron microscopy of Na + ,K + -ATPase in the two E2P states with and without cardiotonic steroids | Kanai R | 2022 Apr 05 |
|
| 35347892 | O‐GlcNAcylation regulates lysophosphatidic acid‐induced cell migration by regulating ERM family proteins | Song M | 2022 Apr 05 |
|
| 35416773 | Author response: Autoinhibition and regulation by phosphoinositides of ATP8B1, a human lipid flippase associated with intrahepatic cholestatic disorders | Dieudonné T | 2022 Apr 05 |
|
| 35377598 | Dichlorophenylpyridine-Based Molecules Inhibit Furin through an Induced-Fit Mechanism | Dahms SO | 2022 Apr 04 |
|
| 35379805 | The engineered CD80 variant fusion therapeutic davoceticept combines checkpoint antagonism with conditional CD28 costimulation for anti-tumor immunity | Maurer MF | 2022 Apr 04 |
|
| 35377815 | Helical self-assembly of a mucin segment suggests an evolutionary origin for von Willebrand factor tubules | Javitt G | 2022 Apr 04 |
|
| 35027402 | Key interactions in the trimolecular complex consisting of the rheumatoid arthritis-associated DRB1*04:01 molecule, the major glycosylated collagen II peptide and the T-cell receptor | Ge C | 2022 Apr |
|
PubAnnotation
| Title | Authors | Source | Publication Date | PubMed ID |
|---|---|---|---|---|
| Dynamic glycosylation of the transcription factor CREB: a potential role in gene regulation. |
|
J Am Chem Soc | 2003 Jun 4 | 12769553 |
| Effect of okadaic acid on O-linked N-acetylglucosamine levels in a neuroblastoma cell line. |
|
Biochim Biophys Acta | 1999 Oct 18 | 10572927 |
| Regulation of a cytosolic and nuclear O-GlcNAc transferase. Role of the tetratricopeptide repeats. |
|
J Biol Chem | 1999 Nov 5 | 10542233 |
| An N-terminal region of Sp1 targets its proteasome-dependent degradation in vitro. |
|
J Biol Chem | 1999 May 21 | 10329728 |
| Vertebrate lens alpha-crystallins are modified by O-linked N-acetylglucosamine. |
|
J Biol Chem | 1992 Jan 5 | 1730617 |
 G14843DJ
G14843DJ
 G68544GH
G68544GH
