Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE G49108TO
G49108TO
Summary
- GlyTouCan ID
-
G49108TO
- IUPAC Condensed
- GlcNAc(b1-
3D Structures
- GLYCAM
- DGlcpNAcb1-OH
Organisms
| Organisms | Evidence |
|---|---|
| Mus musculus (house mouse) | |
| Rattus norvegicus (Norway rat) | |
| Homo sapiens (human) | |
| Drosophila melanogaster (fruit fly) | |
| Streptococcus pneumoniae |
Taxonomic Hierarchy
GlycoGene Database (GGDB)
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| B4GALT5 | (not applicable) |
 G49108TO
G49108TO
|
[beta]-S-pNP |
 G00055MO
G00055MO
|
[beta]-S-pNP | |
| B3GALT2 | UDP-Gal |
 G49108TO
G49108TO
|
Lemieux |
 G00056MO
G00056MO
|
Lemieux | |
| B4GALT2 | UDP-Gal |
 G49108TO
G49108TO
|
Benzyl-[beta] |
 G00055MO
G00055MO
|
Benzyl-[beta] | |
| B4GALT1 | UDP-Gal |
 G49108TO
G49108TO
|
R |
 G00055MO
G00055MO
|
R | |
| B3GALT5 | UDP-Gal |
 G49108TO
G49108TO
|
R |
 G00056MO
G00056MO
|
R |
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| B3GALT5 | UDP-Gal |
 G49108TO
G49108TO
|
R |
 G00056MO
G00056MO
|
R | |
| B3GALT1 | UDP-Gal |
 G49108TO
G49108TO
|
Lemieux |
 G00056MO
G00056MO
|
Lemieux | |
| B4GALT4 | UDP-Gal |
 G49108TO
G49108TO
|
[beta]-1-thio-p-Nitrophenyl |
 G00055MO
G00055MO
|
[beta]-1-thio-p-Nitrophenyl | |
| B4GALNT3 | UDP-GalNAc |
 G49108TO
G49108TO
|
R |
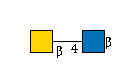 G00057MO
G00057MO
|
R | |
| B3GALNT2 | UDP-GalNAc |
 G49108TO
G49108TO
|
[beta]-Bz |
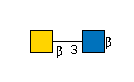 G36387CV
G36387CV
|
[beta]-Bz |
Core Protein
| UniProt ID | Protein Name | Reference | Source |
|---|---|---|---|
| O94762 | ATP-dependent DNA helicase Q5 | ||
| O94763 | Unconventional prefoldin RPB5 interactor 1 | ||
| O94811 | Tubulin polymerization-promoting protein | ||
| O94812 | BAI1-associated protein 3 | ||
| O94817 | Ubiquitin-like protein ATG12 | ||
| O94822 | E3 ubiquitin-protein ligase listerin | ||
| O94823 | Phospholipid-transporting ATPase VB | ||
| O94842 | TOX high mobility group box family member 4 | ||
| O94851 | [F-actin]-monooxygenase MICAL2 | ||
| O94855 | Protein transport protein Sec24D |
GlycoEpitope
- Epitope ID
- EP0004
Pathway
| Pathway Name | Organism |
|---|---|
| Antimicrobial peptides | Drosophila melanogaster |
| Antimicrobial peptides | Rattus norvegicus |
| Antimicrobial peptides | Danio rerio |
| Antimicrobial peptides | Xenopus tropicalis |
| Antimicrobial peptides | Gallus gallus |
| Antimicrobial peptides | Homo sapiens |
| Antimicrobial peptides | Canis familiaris |
| Antimicrobial peptides | Dictyostelium discoideum |
| Antimicrobial peptides | Bos taurus |
| Antimicrobial peptides | Mus musculus |
Sequence Descriptors
- GlycoCT
-
RES 1b:b-dglc-HEX-1:5 2s:n-acetyl LIN 1:1d(2+1)2n
- WURCS
- WURCS=2.0/1,1,0/[a2122h-1b_1-5_2*NCC/3=O]/1/
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 1268028 | Carcinoembryonic antigen: isolation of a sub-fraction with high specific activity | Rogers GT | 1976 Apr |
|
| 176458 | Oligosaccharide moieties of the glycoprotein of vesicular stomatitis virus | Moyer SA | 1976 Apr |
|
| 990335 | [Complete structure of glycopeptides isolated from IgG immunoglobulins of cow colostrum] | Chéron A | 1976 |
|
| 180782 | Some factors in the biliary excretion of estrogens | Watanabe H | 1976 |
|
| 9261 | Studies on insect bacteriolytic enzymes--II. Some physical and enzymatic properties of lysozyme from haemolymph of Galleria mellonella | Powning RF | 1976 |
|
| 827888 | Formation of lipid-bound N-acetylhexosamine derivatives in yeast particulate fractions | Palamarczyk G | 1976 |
|
| 953057 | [Chemistry of urinary mannosides excreted in mannosidosis] | Strecker G | 1976 |
|
| 64195 | Stimulation by GTP of the incorporation of N-acetylglucosamine and mannose into endogenous acceptors of rough endoplasmic reticulum in rat liver | Godelaine D | 1976 |
|
| 134785 | [Biosynthesis of thyroglobulin in an adult lamprey, Lampetra planeri (Bloch)] | Monaco F | 1976 |
|
| 1101894 | Microheterogeneity of the inner core region of yeast manno-protein | Nakajima T | 1975 Sep 16 |
|
PubAnnotation
| Title | Authors | Source | Publication Date | PubMed ID |
|---|---|---|---|---|
| Dynamic glycosylation of the transcription factor CREB: a potential role in gene regulation. |
|
J Am Chem Soc | 2003 Jun 4 | 12769553 |
| Effect of okadaic acid on O-linked N-acetylglucosamine levels in a neuroblastoma cell line. |
|
Biochim Biophys Acta | 1999 Oct 18 | 10572927 |
| Regulation of a cytosolic and nuclear O-GlcNAc transferase. Role of the tetratricopeptide repeats. |
|
J Biol Chem | 1999 Nov 5 | 10542233 |
| An N-terminal region of Sp1 targets its proteasome-dependent degradation in vitro. |
|
J Biol Chem | 1999 May 21 | 10329728 |
| Vertebrate lens alpha-crystallins are modified by O-linked N-acetylglucosamine. |
|
J Biol Chem | 1992 Jan 5 | 1730617 |
 G14843DJ
G14843DJ
 G68544GH
G68544GH
