Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE G49108TO
G49108TO
Summary
- GlyTouCan ID
-
G49108TO
- IUPAC Condensed
- GlcNAc(b1-
3D Structures
- GlycoNAVI
- G49108TO
- GLYCAM
- DGlcpNAcb1-OH
Organisms
| Organisms | Evidence |
|---|---|
| Mus musculus (house mouse) | |
| Rattus norvegicus (Norway rat) | |
| Homo sapiens (human) | |
| Drosophila melanogaster (fruit fly) | |
| Streptococcus pneumoniae |
Taxonomic Hierarchy
GlycoGene Database (GGDB)
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| B4GALT5 | (not applicable) |
 G49108TO
G49108TO
|
[beta]-S-pNP |
 G00055MO
G00055MO
|
[beta]-S-pNP | |
| B3GALT2 | UDP-Gal |
 G49108TO
G49108TO
|
Lemieux |
 G00056MO
G00056MO
|
Lemieux | |
| B4GALT2 | UDP-Gal |
 G49108TO
G49108TO
|
Benzyl-[beta] |
 G00055MO
G00055MO
|
Benzyl-[beta] | |
| B4GALT1 | UDP-Gal |
 G49108TO
G49108TO
|
R |
 G00055MO
G00055MO
|
R | |
| B3GALT5 | UDP-Gal |
 G49108TO
G49108TO
|
R |
 G00056MO
G00056MO
|
R |
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| B4GALNT4 | UDP-GalNAc |
 G49108TO
G49108TO
|
[beta]-Bz |
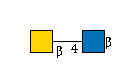 G00057MO
G00057MO
|
[beta]-Bz | |
| B4GALNT3 | UDP-GalNAc |
 G49108TO
G49108TO
|
[beta]-Bz |
 G00057MO
G00057MO
|
[beta]-Bz | |
| B3GALNT2 | UDP-GalNAc |
 G49108TO
G49108TO
|
R |
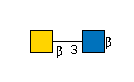 G36387CV
G36387CV
|
R | |
| B4GALNT4 | UDP-GalNAc |
 G49108TO
G49108TO
|
R |
 G00057MO
G00057MO
|
R | |
| B4GALT1 | UDP-Glc |
 G49108TO
G49108TO
|
R |
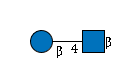 G38884XS
G38884XS
|
R |
Core Protein
GlycoEpitope
- Epitope ID
- EP0004
Pathway
| Pathway Name | Organism |
|---|---|
| Lectin pathway of complement activation | Mus musculus |
| Lectin pathway of complement activation | Bos taurus |
| Lectin pathway of complement activation | Rattus norvegicus |
| Lectin pathway of complement activation | Xenopus tropicalis |
| Lectin pathway of complement activation | Gallus gallus |
| Lectin pathway of complement activation | Homo sapiens |
| Lectin pathway of complement activation | Canis familiaris |
Sequence Descriptors
- GlycoCT
-
RES 1b:b-dglc-HEX-1:5 2s:n-acetyl LIN 1:1d(2+1)2n
- WURCS
- WURCS=2.0/1,1,0/[a2122h-1b_1-5_2*NCC/3=O]/1/
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 36494358 | Structural basis for a human broadly neutralizing influenza A hemagglutinin stem-specific antibody including H17/18 subtypes | Chen Y | 2022 Dec 09 |
|
| 36484366 | <scp>Cryo‐EM</scp> structures of human <scp>ABCA7</scp> provide insights into its phospholipid translocation mechanisms | Le LTM | 2022 Dec 09 |
|
| 36417914 | Structural basis for the severe adverse interaction of sofosbuvir and amiodarone on L-type Cav channels | Yao X | 2022 Dec 08 |
|
| 36477533 | Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides | Yang X | 2022 Dec 07 |
|
| 36478127 | Cytobacillus pseudoceanisediminis sp. nov., A Novel Facultative Methylotrophic Bacterium with High Heavy Metal Resistance Isolated from the Deep Underground Saline Spring | Tarasov K | 2022 Dec 07 |
|
| 36473120 | The role of O-GlcNAcylation in innate immunity and inflammation | Wang Y | 2022 Dec 06 |
|
| 36462113 | Nitratireductor luteus sp. nov. isolated from saline-alkali land | Yang X | 2022 Dec 03 |
|
| 36456728 | Nucleic acid-based scaffold systems and application in enzyme cascade catalysis | Du C | 2022 Dec 02 |
|
| 36309015 | Structural insights into assembly and function of GluN1-2C, GluN1-2A-2C, and GluN1-2D NMDARs | Chou T | 2022 Dec 01 |
|
| 36454514 | Nucleotide sugar transporter SLC35A2 is involved in promoting hepatocellular carcinoma metastasis by regulating cellular glycosylation | Cheng H | 2022 Dec 01 |
|
PubAnnotation
| Title | Authors | Source | Publication Date | PubMed ID |
|---|---|---|---|---|
| Phosphorylation activates master growth regulator DELLA by promoting histone H2A binding at chromatin in Arabidopsis. |
|
Nat Commun | 2024 Sep 3 | 39227587 |
| A protein O-GlcNAc glycosyltransferase regulates the antioxidative response in Yersinia pestis. |
|
Nat Commun | 2024 Aug 16 | 39152136 |
| Polη O-GlcNAcylation governs genome integrity during translesion DNA synthesis. |
|
Nat Commun | 2017 Dec 5 | 29208956 |
| Epithelial Mesenchymal Transition Induces Aberrant Glycosylation through Hexosamine Biosynthetic Pathway Activation. |
|
J Biol Chem | 2016 Jun 17 | 27129262 |
| The making of a sweet modification: structure and function of O-GlcNAc transferase. |
|
J Biol Chem | 2014 Dec 12 | 25336649 |
 G14843DJ
G14843DJ
 G68544GH
G68544GH
