Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE G49108TO
G49108TO
Summary
- GlyTouCan ID
-
G49108TO
- IUPAC Condensed
- GlcNAc(b1-
3D Structures
- GLYCAM
- DGlcpNAcb1-OH
Organisms
| Organisms | Evidence |
|---|---|
| Mus musculus (house mouse) | |
| Rattus norvegicus (Norway rat) | |
| Homo sapiens (human) | |
| Drosophila melanogaster (fruit fly) | |
| Streptococcus pneumoniae |
Taxonomic Hierarchy
GlycoGene Database (GGDB)
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| B4GALT5 | (not applicable) |
 G49108TO
G49108TO
|
[beta]-S-pNP |
 G00055MO
G00055MO
|
[beta]-S-pNP | |
| B3GALT2 | UDP-Gal |
 G49108TO
G49108TO
|
Lemieux |
 G00056MO
G00056MO
|
Lemieux | |
| B4GALT2 | UDP-Gal |
 G49108TO
G49108TO
|
Benzyl-[beta] |
 G00055MO
G00055MO
|
Benzyl-[beta] | |
| B4GALT1 | UDP-Gal |
 G49108TO
G49108TO
|
R |
 G00055MO
G00055MO
|
R | |
| B3GALT5 | UDP-Gal |
 G49108TO
G49108TO
|
R |
 G00056MO
G00056MO
|
R |
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| B4GALNT4 | UDP-GalNAc |
 G49108TO
G49108TO
|
[beta]-Bz |
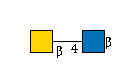 G00057MO
G00057MO
|
[beta]-Bz | |
| B4GALNT3 | UDP-GalNAc |
 G49108TO
G49108TO
|
[beta]-Bz |
 G00057MO
G00057MO
|
[beta]-Bz | |
| B3GALNT2 | UDP-GalNAc |
 G49108TO
G49108TO
|
R |
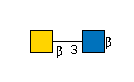 G36387CV
G36387CV
|
R | |
| B4GALNT4 | UDP-GalNAc |
 G49108TO
G49108TO
|
R |
 G00057MO
G00057MO
|
R | |
| B4GALT1 | UDP-Glc |
 G49108TO
G49108TO
|
R |
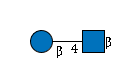 G38884XS
G38884XS
|
R |
Core Protein
GlycoEpitope
- Epitope ID
- EP0004
Pathway
| Pathway Name | Organism |
|---|---|
| Antimicrobial peptides | Sus scrofa |
| E.coli O10 | Escherichia coli |
| E.coli O103 | Escherichia coli |
| E.coli O105 | Escherichia coli |
| E.coli O109 | Escherichia coli |
| E.coli O110 | Escherichia coli |
| E.coli O111 | Escherichia coli |
| E.coli O112ab | Escherichia coli |
| E.coli O113 | Escherichia coli |
| E.coli O115 | Escherichia coli |
Sequence Descriptors
- GlycoCT
-
RES 1b:b-dglc-HEX-1:5 2s:n-acetyl LIN 1:1d(2+1)2n
- WURCS
- WURCS=2.0/1,1,0/[a2122h-1b_1-5_2*NCC/3=O]/1/
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 3731182 | The effect of a 'bisecting' N-acetylglucosaminyl group on the binding of biantennary, complex oligosaccharides to concanavalin A, Phaseolus vulgaris erythroagglutinin (E-PHA), and Ricinus communis agglutinin (RCA-120) immobilized on agarose | Narasimhan S | 1986 Jun 01 |
|
| 3726811 | Study on the endogenous lectins of the human platelet plasma membrane | Scurei C | 1986 Jul 01 |
|
| 3519750 | Ultrastructural localization of glycoconjugates in the fungus Ascocalyx abietina, the Scleroderris canker agent of conifers, using lectin-gold complexes. | Benhamou N | 1986 Jul |
|
| 3708596 | Similarities in glycosylation of human neuroblastoma tumors and cell lines | Woodbury RA | 1986 Jul |
|
| 3014508 | Cloning and sequencing of cDNA of bovine N-acetylglucosamine (beta 1-4)galactosyltransferase. | Narimatsu H | 1986 Jul |
|
| 2869841 | Discrimination of gamma-glutamyltranspeptidase from normal and carcinomatous pancreas | Yamaguchi N | 1986 Jan 30 |
|
| 3949914 | Simultaneous determination of N-acetylglucosamine, N-acetylgalactosamine, N-acetylglucosaminitol and N-acetylgalactosaminitol by gas—liquid chromatography | Mawhinney TP | 1986 Jan 03 |
|
| 2934286 | New Diabetogenic Streptozocin Analogue, 3-O-Methyl-2-{[(methylnitrosoamino) carbonyl]amino}-D-glucopyranose: Evidence for a Glucose Recognition Site on Pancreatic B-Cells | Kawada J | 1986 Jan 01 |
|
| 3519852 | A Candida albicans Mutant Impaired in the Utilization of N-Acetylglucosamine | Corner BE | 1986 Jan 01 |
|
| 2877789 | The pharate adult clasper as a tool for measuring chitin synthesis and for identifying new chitin synthesis inhibitors | Gelman DB | 1986 Jan |
|
PubAnnotation
| Title | Authors | Source | Publication Date | PubMed ID |
|---|---|---|---|---|
| Dynamic glycosylation of the transcription factor CREB: a potential role in gene regulation. |
|
J Am Chem Soc | 2003 Jun 4 | 12769553 |
| Effect of okadaic acid on O-linked N-acetylglucosamine levels in a neuroblastoma cell line. |
|
Biochim Biophys Acta | 1999 Oct 18 | 10572927 |
| Regulation of a cytosolic and nuclear O-GlcNAc transferase. Role of the tetratricopeptide repeats. |
|
J Biol Chem | 1999 Nov 5 | 10542233 |
| An N-terminal region of Sp1 targets its proteasome-dependent degradation in vitro. |
|
J Biol Chem | 1999 May 21 | 10329728 |
| Vertebrate lens alpha-crystallins are modified by O-linked N-acetylglucosamine. |
|
J Biol Chem | 1992 Jan 5 | 1730617 |
 G14843DJ
G14843DJ
 G68544GH
G68544GH
