Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE G49108TO
G49108TO
Summary
- GlyTouCan ID
-
G49108TO
- IUPAC Condensed
- GlcNAc(b1-
External Links
- GlyConnect structure
- GlyGen
- ChEBI
- PubChem Compound
- PubChem Substance
- The Human Metabolome Database
- CarbBank
- 9 3780 3968 4708 25653 28493 29021 29527 29549 31775 32028 32903 33539 36048 36270 37035 37525 37675 37677 37678 39452 39454 39456 39609 39611 39613 39614 40149 40150 40151 40355 40591 40600 42059 42207 42220 43184 43893 43925 43926 43931 43932 43941 44395 44396 44397 44549 46453 47268 47468 47537 47603 47796 48630 48812 48813 49006 49732 50589 50736
3D Structures
- GLYCAM
- DGlcpNAcb1-OH
Organisms
| Organisms | Evidence |
|---|---|
| Streptococcus pneumoniae | |
| Mus musculus (house mouse) | |
| Saccharomyces cerevisiae (brewer's yeast) | |
| Sus scrofa (pig) | |
| Rattus norvegicus (Norway rat) |
Taxonomic Hierarchy
GlycoGene Database (GGDB)
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| B4GALT5 | (not applicable) |
 G49108TO
G49108TO
|
[beta]-S-pNP |
 G00055MO
G00055MO
|
[beta]-S-pNP | |
| B4GALT1 | UDP-Gal |
 G49108TO
G49108TO
|
R |
 G00055MO
G00055MO
|
R | |
| B4GALT2 | UDP-Gal |
 G49108TO
G49108TO
|
Benzyl-[beta] |
 G00055MO
G00055MO
|
Benzyl-[beta] | |
| B3GALT2 | UDP-Gal |
 G49108TO
G49108TO
|
Lemieux |
 G00056MO
G00056MO
|
Lemieux | |
| B3GALT5 | UDP-Gal |
 G49108TO
G49108TO
|
R |
 G00056MO
G00056MO
|
R |
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| B4GALNT3 | UDP-GalNAc |
 G49108TO
G49108TO
|
[beta]-Bz |
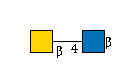 G00057MO
G00057MO
|
[beta]-Bz | |
| B3GALNT2 | UDP-GalNAc |
 G49108TO
G49108TO
|
[beta]-Bz |
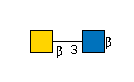 G36387CV
G36387CV
|
[beta]-Bz | |
| B4GALNT4 | UDP-GalNAc |
 G49108TO
G49108TO
|
[beta]-Bz |
 G00057MO
G00057MO
|
[beta]-Bz | |
| B3GALNT2 | UDP-GalNAc |
 G49108TO
G49108TO
|
R |
 G36387CV
G36387CV
|
R | |
| B4GALT1 | UDP-Glc |
 G49108TO
G49108TO
|
R |
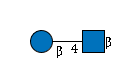 G38884XS
G38884XS
|
R |
Core Protein
| UniProt ID | Protein Name | Reference | Source |
|---|---|---|---|
| Q9NZN5 | Rho guanine nucleotide exchange factor 12 | ||
| Q9NZN8 | CCR4-NOT transcription complex subunit 2 | ||
| Q9NZQ3 | NCK-interacting protein with SH3 domain | ||
| Q9NZR1 | Tropomodulin-2 | ||
| Q9NZR2 | Low-density lipoprotein receptor-related protein 1B | ||
| Q9NZU1 | Leucine-rich repeat transmembrane protein FLRT1 | ||
| Q9NZV1 | Cysteine-rich motor neuron 1 protein | ||
| Q9NZV5 | Selenoprotein N | ||
| Q9NZZ3 | Charged multivesicular body protein 5 | ||
| Q9P016 | Thymocyte nuclear protein 1 |
GlycoEpitope
- Epitope ID
- EP0004
Pathway
| Pathway Name | Organism |
|---|---|
| Antimicrobial peptides | Sus scrofa |
| Antimicrobial peptides | Rattus norvegicus |
| Antimicrobial peptides | Dictyostelium discoideum |
| Antimicrobial peptides | Bos taurus |
| Antimicrobial peptides | Canis familiaris |
| Antimicrobial peptides | Mus musculus |
| Antimicrobial peptides | Gallus gallus |
| Antimicrobial peptides | Homo sapiens |
| Antimicrobial peptides | Drosophila melanogaster |
| Antimicrobial peptides | Xenopus tropicalis |
Sequence Descriptors
- GlycoCT
-
RES 1b:b-dglc-HEX-1:5 2s:n-acetyl LIN 1:1d(2+1)2n
- WURCS
- WURCS=2.0/1,1,0/[a2122h-1b_1-5_2*NCC/3=O]/1/
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 38512671 | The O-GlcNAc Modification of Recombinant Tau Protein and Characterization of the O-GlcNAc Pattern for Functional Study | El Hajjar L | 2024 |
|
| 37737979 | Characterization of T-Cell Epitopes in Food Allergens by Bioinformatic Tools | He S | 2024 |
|
| 38963494 | The Prothrombin-Prothrombinase Interaction | Stojanovski BM | 2024 |
|
| 38995536 | Integrating HexNAcQuest with Glycoproteomics Data Analysis Software to Distinguish HexNAc Isomers on Peptides | Hou C | 2024 |
|
| 39267774 | Bisecting GlcNAc modification reverses the chemoresistance via attenuating the function of P-gp | Tan Z | 2024 |
|
| 38385070 | A novel CSN5/CRT O-GlcNAc/ER stress regulatory axis in platinum resistance of epithelial ovarian cancer | Yan T | 2024 |
|
| 38347423 | Bacterial Enzyme Assay for Mucin Glycan Degradation | Katoh T | 2024 |
|
| 37033243 | O-GlcNAcylation-induced GSK-3β activation deteriorates pressure overload-induced heart failure via lack of compensatory cardiac hypertrophy in mice | Matsuno M | 2023 |
|
| 38192280 | O-GlcNAc regulates the mitochondrial integrated stress response by regulating ATF4 | Alghusen IM | 2023 |
|
| 38097051 | Pharmacological activation of nuclear factor erythroid 2-related factor-2 prevents hyperglycemia-induced renal oxidative damage: Possible involvement of O-GlcNAcylation | Costa RM | 2023-12-12 |
|
 G14843DJ
G14843DJ
 G68544GH
G68544GH
