Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE G49108TO
G49108TO
Summary
- GlyTouCan ID
-
G49108TO
- IUPAC Condensed
- GlcNAc(b1-
External Links
- GlyConnect structure
- GlyGen
- ChEBI
- PubChem Compound
- PubChem Substance
- The Human Metabolome Database
- CarbBank
- 9 3780 3968 4708 25653 28493 29021 29527 29549 31775 32028 32903 33539 36048 36270 37035 37525 37675 37677 37678 39452 39454 39456 39609 39611 39613 39614 40149 40150 40151 40355 40591 40600 42059 42207 42220 43184 43893 43925 43926 43931 43932 43941 44395 44396 44397 44549 46453 47268 47468 47537 47603 47796 48630 48812 48813 49006 49732 50589 50736
3D Structures
- GLYCAM
- DGlcpNAcb1-OH
Organisms
| Organisms | Evidence |
|---|---|
| Streptococcus pneumoniae | |
| Mus musculus (house mouse) | |
| Saccharomyces cerevisiae (brewer's yeast) | |
| Sus scrofa (pig) | |
| Rattus norvegicus (Norway rat) |
Taxonomic Hierarchy
GlycoGene Database (GGDB)
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| B4GALT5 | (not applicable) |
 G49108TO
G49108TO
|
[beta]-S-pNP |
 G00055MO
G00055MO
|
[beta]-S-pNP | |
| B4GALT1 | UDP-Gal |
 G49108TO
G49108TO
|
R |
 G00055MO
G00055MO
|
R | |
| B4GALT2 | UDP-Gal |
 G49108TO
G49108TO
|
Benzyl-[beta] |
 G00055MO
G00055MO
|
Benzyl-[beta] | |
| B3GALT2 | UDP-Gal |
 G49108TO
G49108TO
|
Lemieux |
 G00056MO
G00056MO
|
Lemieux | |
| B3GALT5 | UDP-Gal |
 G49108TO
G49108TO
|
R |
 G00056MO
G00056MO
|
R |
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| B4GALNT3 | UDP-GalNAc |
 G49108TO
G49108TO
|
[beta]-Bz |
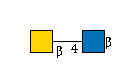 G00057MO
G00057MO
|
[beta]-Bz | |
| B3GALNT2 | UDP-GalNAc |
 G49108TO
G49108TO
|
[beta]-Bz |
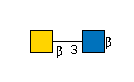 G36387CV
G36387CV
|
[beta]-Bz | |
| B4GALNT4 | UDP-GalNAc |
 G49108TO
G49108TO
|
[beta]-Bz |
 G00057MO
G00057MO
|
[beta]-Bz | |
| B3GALNT2 | UDP-GalNAc |
 G49108TO
G49108TO
|
R |
 G36387CV
G36387CV
|
R | |
| B4GALT1 | UDP-Glc |
 G49108TO
G49108TO
|
R |
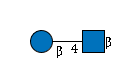 G38884XS
G38884XS
|
R |
Core Protein
| UniProt ID | Protein Name | Reference | Source |
|---|---|---|---|
| O70511 | Ankyrin-3 | ||
| O70546 | Lysine-specific demethylase 6A | ||
| O70551 | SRSF protein kinase 1 | ||
| O70585 | Dystrobrevin beta | ||
| O70591 | Prefoldin subunit 2 | ||
| O75027 | Iron-sulfur clusters transporter ABCB7, mitochondrial | ||
| P08F94 | Fibrocystin | ||
| O75030 | Microphthalmia-associated transcription factor | ||
| O75037 | Kinesin-like protein KIF21B | ||
| O75038 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase eta-2 |
GlycoEpitope
- Epitope ID
- EP0004
Pathway
| Pathway Name | Organism |
|---|---|
| Antimicrobial peptides | Sus scrofa |
| Antimicrobial peptides | Rattus norvegicus |
| Antimicrobial peptides | Dictyostelium discoideum |
| Antimicrobial peptides | Bos taurus |
| Antimicrobial peptides | Canis familiaris |
| Antimicrobial peptides | Mus musculus |
| Antimicrobial peptides | Gallus gallus |
| Antimicrobial peptides | Homo sapiens |
| Antimicrobial peptides | Drosophila melanogaster |
| Antimicrobial peptides | Xenopus tropicalis |
Sequence Descriptors
- GlycoCT
-
RES 1b:b-dglc-HEX-1:5 2s:n-acetyl LIN 1:1d(2+1)2n
- WURCS
- WURCS=2.0/1,1,0/[a2122h-1b_1-5_2*NCC/3=O]/1/
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 37137348 | Identification and functional characterisation of N-acetylglucosamine kinase from Helicoverpa armigera divulge its potential role in growth and development via UDP-GlcNAc salvage pathway | Das J | 2023 Jul 01 |
|
| 37028529 | Production of N-acetylglucosamine from carbon dioxide by engineering Cupriavidus necator H16 | Wang X | 2023 Jul |
|
| 36586406 | A delicate balance between antibody evasion and ACE2 affinity for Omicron BA.2.75 | Huo J | 2023 Jan 31 |
|
| 36720854 | Mechanism of hormone and allosteric agonist mediated activation of follicle stimulating hormone receptor | Duan J | 2023 Jan 31 |
|
| 36720859 | Molecular insights into the gating mechanisms of voltage-gated calcium channel CaV2.3 | Gao Y | 2023 Jan 31 |
|
| 36640338 | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein | Saville JW | 2023 Jan 31 |
|
| 36511698 | Characterization of a Novel CD4 Mimetic Compound YIR-821 against HIV-1 Clinical Isolates | Matsumoto K | 2023 Jan 31 |
|
| 36776401 | Chemical biology tools to interrogate the roles of O-GlcNAc in immunity | Saha A | 2023 Jan 26 |
|
| 36714014 | Design of hACE2-based small peptide inhibitors against spike protein of SARS-CoV-2: a computational approach | Dhingra N | 2023 Jan 25 |
|
| 36708708 | Structure of the Inmazeb cocktail and resistance to escape against Ebola virus | Rayaprolu V | 2023 Jan 25 |
|
 G14843DJ
G14843DJ
 G68544GH
G68544GH
