Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE G60230HH
G60230HH
Summary
- GlyTouCan ID
-
G60230HH
- IUPAC Condensed
- Man(a1-2)Man(a1-2)Man(a1-3)[Man(a1-2)Man(a1-3)[Man(a1-2)Man(a1-6)]Man(a1-6)]Man(b1-4)GlcNAc(b1-4)GlcNAc(b1-
- Motifs
- N-Glycan high mannose
- Subsumption Level
- Fully-defined saccharide
- Monoisotopic Mass
- 1882.64
3D Structures
Organisms
| Organisms | Evidence |
|---|---|
| Saccharomyces cerevisiae (brewer's yeast) | |
| Mus musculus (house mouse) | |
| Drosophila melanogaster (fruit fly) | |
| Homo sapiens (human) | |
| Phaseolus vulgaris |
Taxonomic Hierarchy
GlycoGene Database (GGDB)
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| UGGT1 (UGCGL1) | UDP-Glc |
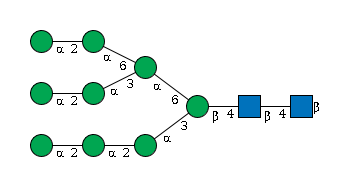 G60230HH
G60230HH
|
MTX |
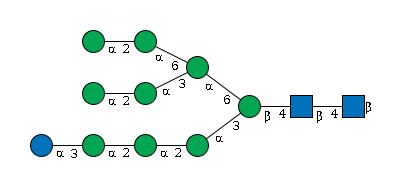 G19958IL
G19958IL
|
MTX | |
| UGGT2 (UGCGL2) | UDP-Glc |
 G60230HH
G60230HH
|
MTX |
 G19958IL
G19958IL
|
MTX |
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| UGGT1 (UGCGL1) | UDP-Glc |
 G60230HH
G60230HH
|
MTX |
 G19958IL
G19958IL
|
MTX | |
| UGGT2 (UGCGL2) | UDP-Glc |
 G60230HH
G60230HH
|
MTX |
 G19958IL
G19958IL
|
MTX |
KEGG BRITE Database
GALAXY
Glucose Units
- ODS
- 5.2
- Amide
- 9.7
Glycans containing this structure in GALAXY
Core Protein
| UniProt ID | Protein Name | Reference | Source |
|---|---|---|---|
| P35439 | Glutamate receptor ionotropic, NMDA 1 | ||
| P54290 | Voltage-dependent calcium channel subunit alpha-2/delta-1 | ||
| P56526 | Alpha-glucosidase | ||
| P56817 | Beta-secretase 1 | ||
| P61823 | Ribonuclease pancreatic | ||
| P83180 | Hemocyanin B chain | ||
| P97846 | Contactin-associated protein 1 | ||
| Q00960 | Glutamate receptor ionotropic, NMDA 2B | ||
| Q02223 | Tumor necrosis factor receptor superfamily member 17 | ||
| Q08169 | Hyaluronidase |
Sequence Descriptors
- GlycoCT
-
RES 1b:b-dglc-HEX-1:5 2s:n-acetyl 3b:b-dglc-HEX-1:5 4s:n-acetyl 5b:b-dman-HEX-1:5 6b:a-dman-HEX-1:5 7b:a-dman-HEX-1:5 8b:a-dman-HEX-1:5 9b:a-dman-HEX-1:5 10b:a-dman-HEX-1:5 11b:a-dman-HEX-1:5 12b:a-dman-HEX-1:5 13b:a-dman-HEX-1:5 LIN 1:1d(2+1)2n 2:1o(4+1)3d 3:3d(2+1)4n 4:3o(4+1)5d 5:5o(3+1)6d 6:6o(2+1)7d 7:7o(2+1)8d 8:5o(6+1)9d 9:9o(3+1)10d 10:10o(2+1)11d 11:9o(6+1)12d 12:12o(2+1)13d
- WURCS
- WURCS=2.0/3,11,10/[a2122h-1b_1-5_2*NCC/3=O][a1122h-1b_1-5][a1122h-1a_1-5]/1-1-2-3-3-3-3-3-3-3-3/a4-b1_b4-c1_c3-d1_c6-g1_d2-e1_e2-f1_g3-h1_g6-j1_h2-i1_j2-k1
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 35022400 | Mammalian brain glycoproteins exhibit diminished glycan complexity compared to other tissues | Williams SE | 2022 Jan 12 |
|
| 33242084 | Post-natal developmental changes in the composition of the rat neocortical N-glycome | Klarić Thomas S | 2021-06-03 |
|
| 33737529 | Toward robust N-glycomics of various tissue samples that may contain glycans with unknown or unexpected structures | Suzuki Noriko | 2021-03-18 |
|
| 34834161 | Glycan Profile Analysis of Engineered Trastuzumab with Rationally Added Glycosylation Sequons Presents Significantly Increased Glycan Complexity | Cruz E | 2021 Oct 20 |
|
| 33993882 | Complete spatial characterisation of N-glycosylation upon striatal neuroinflammation in the rodent brain | Rebelo AL | 2021 May 16 |
|
| 34106099 | N-Glycosylation in isolated rat nerve terminals | Matthies I | 2021 Aug 1 |
|
| 33139572 | Spatial and temporal diversity of glycome expression in mammalian brain | Lee Jua | 2020-11-17 |
|
| 29549260 | HIV envelope V3 region mimic embodies key features of a broadly neutralizing antibody lineage epitope | Fera D | 2018 Mar 16 |
|
| 25802287 | A Microarray-Matrix-assisted Laser Desorption/Ionization-Mass Spectrometry Approach for Site-specific Protein N-glycosylation Analysis, as Demonstrated for Human Serum Immunoglobulin M (IgM) | Pabst M | 2015 Jun |
|
| 26510530 | A Method for Comprehensive Glycosite-Mapping and Direct Quantitation of Serum Glycoproteins | Hong Q | 2015 Dec 4 |
|
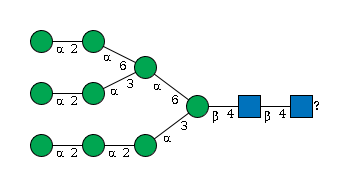 G92042VQ
G92042VQ
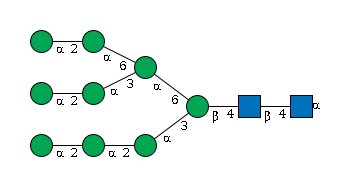 G39578OU
G39578OU
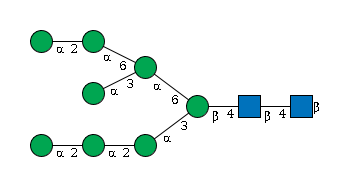 G40702WU
G40702WU
