Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE G71142DF
G71142DF
Summary
- GlyTouCan ID
-
G71142DF
- IUPAC Condensed
- Glc(b1-
- Motifs
- Glucosylceramide
External Links
- GlyGen
- ChEBI
- PubChem Compound
- PubChem Substance
- The Human Metabolome Database
3D Structures
- GlycoNAVI
- G71142DF
- GLYCAM
- DGlcpb1-OH
Organisms
| Organisms | Evidence |
|---|---|
| Mus musculus (house mouse) | |
| Clostridium | |
| Chloroflexus aurantiacus | |
| Homo sapiens (human) | |
| Glycine max (soybean) |
Taxonomic Hierarchy
GlycoGene Database (GGDB)
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| B4GALT5 | (not applicable) |
 G71142DF
G71142DF
|
Ceramide |
 G84224TW
G84224TW
|
Ceramide | |
| B4GALT6 | UDP-Gal |
 G71142DF
G71142DF
|
Cer |
 G84224TW
G84224TW
|
Cer | |
| GXYLT1 | UDP-Xyl |
 G71142DF
G71142DF
|
Ser |
 G01629YL
G01629YL
|
Ser | |
| GXYLT2 | UDP-Xyl |
 G71142DF
G71142DF
|
Ser |
 G01629YL
G01629YL
|
Ser |
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| B4GALT5 | (not applicable) |
 G71142DF
G71142DF
|
Ceramide |
 G84224TW
G84224TW
|
Ceramide | |
| B3GALT2 | UDP-Gal |
 G71142DF
G71142DF
|
Cer |
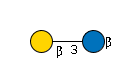 G31761VE
G31761VE
|
Cer | |
| B4GALT1 | UDP-Gal |
 G71142DF
G71142DF
|
R |
 G84224TW
G84224TW
|
R | |
| B4GALT6 | UDP-Gal |
 G71142DF
G71142DF
|
 G84224TW
G84224TW
|
|||
| B4GALT6 | UDP-Gal |
 G71142DF
G71142DF
|
Cer |
 G84224TW
G84224TW
|
Cer |
KEGG BRITE Database
- KEGG ID
- Enzyme
- Reactions
Pathway
| Pathway Name | Organism |
|---|---|
| E.coli O102 | Escherichia coli |
| E.coli O103 | Escherichia coli |
| E.coli O130 | Escherichia coli |
| E.coli O134 | Escherichia coli |
| E.coli O137 | Escherichia coli |
| E.coli O139 | Escherichia coli |
| E.coli O141 | Escherichia coli |
| E.coli O150 | Escherichia coli |
| E.coli O152 | Escherichia coli |
| E.coli O157 | Escherichia coli |
Sequence Descriptors
- GlycoCT
-
RES 1b:b-dglc-HEX-1:5
- WURCS
- WURCS=2.0/1,1,0/[a2122h-1b_1-5]/1/
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 38907752 | Machine learning–driven SERS analysis platform for rapid and accurate detection of precancerous lesions of gastric cancer | Cao D | 2024 Jun 22 |
|
| 38877163 | Identification of mungbean yellow mosaic India virus and susceptibility-related metabolites in the apoplast of mung bean leaves | Dhobale KV | 2024 Jun 14 |
|
| 38871719 | Comprehensive multi-omics analysis of breast cancer reveals distinct long-term prognostic subtypes | Sharma A | 2024 Jun 13 |
|
| 38219565 | Impact of washing and freezing on nutritional composition, bioactive compounds, antioxidant activity and microstructure of mango peels | Marçal S | 2024 Jun 01 |
|
| 39282197 | Production, Purification, and Characterization of a Novel Exopolysaccharide from Probiotic Lactobacillus amylovorus: MTCC 8129 | Murugu J | 2024 Jul 05 |
|
| 38954242 | Evaluation of co-culture of cellulolytic fungi for enhanced cellulase and xylanase activity and saccharification of untreated lignocellulosic material | Kathirgamanathan M | 2024 Jul 02 |
|
| 38279235 | Abscisic Acid Affects Phenolic Acid Content to Increase Tolerance to UV-B Stress in Rhododendron chrysanthum Pall. | Zhou X | 2024 Jan 19 |
|
| 38183486 | The application of bacteria-derived dehydrogenases and oxidases in the synthesis of gold nanoparticles | Martinaga L | 2024 Jan 06 |
|
| 38180643 | A Broad-Spectrum α-Glucosidase of Glycoside Hydrolase Family 13 from Marinovum sp., a Member of the Roseobacter Clade | Li J | 2024 Jan 05 |
|
| 38393628 | Limosilactobacillus fermentum Strains as Novel Probiotic Candidates to Promote Host Health Benefits and Development of Biotherapeutics: A Comprehensive Review | de Luna Freire MO | 2024 Feb 23 |
|
 G80753ZP
G80753ZP
 G58161NS
G58161NS
 G24620AI
G24620AI
 G98256YA
G98256YA
 G72079JG
G72079JG
