Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE G71142DF
G71142DF
Summary
- GlyTouCan ID
-
G71142DF
- IUPAC Condensed
- Glc(b1-
- Motifs
- Glucosylceramide
External Links
- GlyGen
- ChEBI
- PubChem Compound
- PubChem Substance
- The Human Metabolome Database
3D Structures
- GlycoNAVI
- G71142DF
- GLYCAM
- DGlcpb1-OH
Organisms
| Organisms | Evidence |
|---|---|
| Mus musculus (house mouse) | |
| Clostridium | |
| Chloroflexus aurantiacus | |
| Homo sapiens (human) | |
| Glycine max (soybean) |
Taxonomic Hierarchy
GlycoGene Database (GGDB)
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| B4GALT5 | (not applicable) |
 G71142DF
G71142DF
|
Ceramide |
 G84224TW
G84224TW
|
Ceramide | |
| B4GALT6 | UDP-Gal |
 G71142DF
G71142DF
|
Cer |
 G84224TW
G84224TW
|
Cer | |
| GXYLT1 | UDP-Xyl |
 G71142DF
G71142DF
|
Ser |
 G01629YL
G01629YL
|
Ser | |
| GXYLT2 | UDP-Xyl |
 G71142DF
G71142DF
|
Ser |
 G01629YL
G01629YL
|
Ser |
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| B4GALT5 | (not applicable) |
 G71142DF
G71142DF
|
Ceramide |
 G84224TW
G84224TW
|
Ceramide | |
| B3GALT2 | UDP-Gal |
 G71142DF
G71142DF
|
Cer |
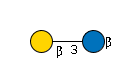 G31761VE
G31761VE
|
Cer | |
| B4GALT1 | UDP-Gal |
 G71142DF
G71142DF
|
R |
 G84224TW
G84224TW
|
R | |
| B4GALT6 | UDP-Gal |
 G71142DF
G71142DF
|
 G84224TW
G84224TW
|
|||
| B4GALT6 | UDP-Gal |
 G71142DF
G71142DF
|
Cer |
 G84224TW
G84224TW
|
Cer |
KEGG BRITE Database
- KEGG ID
- Enzyme
- Reactions
Pathway
| Pathway Name | Organism |
|---|---|
| E.coli O102 | Escherichia coli |
| E.coli O103 | Escherichia coli |
| E.coli O130 | Escherichia coli |
| E.coli O134 | Escherichia coli |
| E.coli O137 | Escherichia coli |
| E.coli O139 | Escherichia coli |
| E.coli O141 | Escherichia coli |
| E.coli O150 | Escherichia coli |
| E.coli O152 | Escherichia coli |
| E.coli O157 | Escherichia coli |
Sequence Descriptors
- GlycoCT
-
RES 1b:b-dglc-HEX-1:5
- WURCS
- WURCS=2.0/1,1,0/[a2122h-1b_1-5]/1/
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 38396285 | Fungal polysaccharides from Inonotus obliquus are agonists for Toll-like receptors and induce macrophage anti-cancer activity | Wold CW | 2024 Feb 23 |
|
| 38342915 | An efficient CRISPR/Cas9 genome editing system based on a multiple sgRNA processing platform in Trichoderma reesei for strain improvement and enzyme production | Zhang J | 2024 Feb 11 |
|
| 39661226 | Curcumin mediates glutathione depletion via metal–organic framework nanocarriers to enhance cisplatin chemosensitivity on esophageal cancer | Sun Y | 2024 Dec 11 |
|
| 39653770 | Synthetic molecular cage receptors for carbohydrate recognition | Wu B | 2024 Dec 09 |
|
| 39621117 | Behavior and mechanism of element dissolution from albite by mannose: experimental and theorical study | Zhang L | 2024 Dec 02 |
|
| 39207679 | Glucose Oxidase Coupling with Pistol-Like DNAzyme Based Colorimetric Assay for Sensitive Glucose Detection in Tears and Saliva | Fan J | 2024 Aug 29 |
|
| 39917349 | 16S metagenomics and metabolomics unveil the microbial compositions and metabolite profiles in Dahi, a traditional Indian fermented milk product prepared by the backslopping method | Tigga A | 2024 Aug 22 |
|
| 39448541 | Author response for 'Crystal structures of Aspergillus oryzae exo‐β‐(1,3)‐glucanase reveal insights into oligosaccharide binding, recognition, and hydrolysis' | Barnava Banerjee | 2024 Aug 22 |
|
| 39158815 | Eleven new glycosidic acid methyl esters from the crude resin glycoside fraction of Ipomoea alba seeds | Misuda N | 2024 Aug 19 |
|
| 39145831 | Aromatic residues in N-terminal domain of archaeal trehalase affect the folding and activity of catalytic domain | Sakoh T | 2024 Aug 15 |
|
 G80753ZP
G80753ZP
 G58161NS
G58161NS
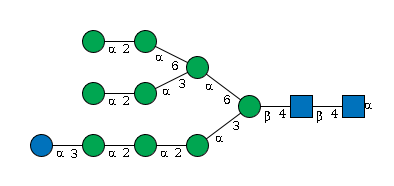 G24620AI
G24620AI
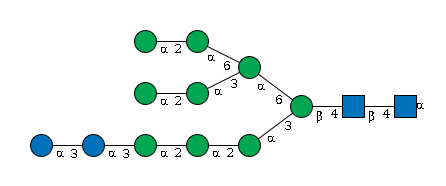 G98256YA
G98256YA
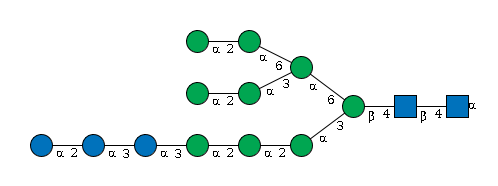 G72079JG
G72079JG
