Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE G96881BQ
G96881BQ
External Links
- GlyConnect structure
- GlyGen
- ChEBI
- PubChem Compound
- PubChem Substance
- The Human Metabolome Database
- CarbBank
3D Structures
- GLYCAM
- LFucpa1-OH
Organisms
Taxonomic Hierarchy
GlycoGene Database (GGDB)
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| B3GLCT | UDP-Glc |
 G96881BQ
G96881BQ
|
rat F-spondin TSR4 |
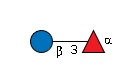 G36855WW
G36855WW
|
rat F-spondin TSR4 | |
| B3GLCT | UDP-Glc |
 G96881BQ
G96881BQ
|
[alpha]-pNp |
 G36855WW
G36855WW
|
[alpha]-pNp | |
| MFNG | UDP-GlcNAc |
 G96881BQ
G96881BQ
|
-O-EGF |
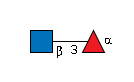 G26238GL
G26238GL
|
-O-EGF | |
| MFNG | UDP-GlcNAc |
 G96881BQ
G96881BQ
|
-pNP |
 G26238GL
G26238GL
|
-pNP | |
| LFNG | UDP-GlcNAc |
 G96881BQ
G96881BQ
|
[alpha]-pNp |
 G26238GL
G26238GL
|
[alpha]-pNp |
KEGG BRITE Database
- KEGG ID
Core Protein
| UniProt ID | Protein Name | Reference | Source |
|---|---|---|---|
| P00740 | Coagulation factor IX | ||
| P00749 | Urokinase-type plasminogen activator | ||
| P00750 | Tissue-type plasminogen activator | ||
| P07996 | Thrombospondin-1 | ||
| P08709 | Coagulation factor VII | ||
| P14328 | Spore coat protein SP96 | ||
| Q09163 | Protein delta homolog 1 | ||
| Q76LX8 | A disintegrin and metalloproteinase with thrombospondin motifs 13 |
Pathway
| Pathway Name | Organism |
|---|---|
| E.coli O11 | Escherichia coli |
| E.coli O125ab | Escherichia coli |
| E.coli O126H27 | Escherichia coli |
| E.coli O127 | Escherichia coli |
| E.coli O128ab | Escherichia coli |
| E.coli O128ab-H27 | Escherichia coli |
| E.coli O128ac | Escherichia coli |
| E.coli O156 | Escherichia coli |
| E.coli O157 | Escherichia coli |
| E.coli O159 | Escherichia coli |
Sequence Descriptors
- GlycoCT
-
RES 1b:a-lgal-HEX-1:5|6:d
- WURCS
- WURCS=2.0/1,1,0/[a1221m-1a_1-5]/1/
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 39256596 | Terminal α1,2-fucosylation of glycosphingolipids by FUT1 is a key regulator in early cell-fate decisions | Chen S | 2024 Sep 10 |
|
| 39464521 | Reappraisal of different agro-industrial waste for the optimization of cellulase production from Aspergillus niger ITV02 in a liquid medium using a Box–Benkhen design | Miranda-Sosa A | 2024 Oct 23 |
|
| 38771321 | Synthesis of fucosyllactose using α-L-fucosidases GH29 from infant gut microbial metagenome | Moya-Gonzálvez EM | 2024 May 21 |
|
| 38890439 | Exploring the sequence-function space of microbial fucosidases | Martínez Gascueña A | 2024 Jun 18 |
|
| 39066828 | Decoding the TAome and computational insights into parDE toxin-antitoxin systems in Pseudomonas aeruginosa | Gupta N | 2024 Jul 27 |
|
| 38985762 | An RNA aptamer exploits exosite-dependent allostery to achieve specific inhibition of coagulation factor IXa | Kolyadko VN | 2024 Jul 10 |
|
| 38182984 | Comparative transcriptome analysis revealed that auxin and cell wall biosynthesis play important roles in the formation of hollow hearts in cucumber | Li J | 2024 Jan 05 |
|
| 38668917 | Genome Analysis of Multiple Polysaccharide-Degrading Bacterium Microbulbifer thermotolerans HB226069: Determination of Alginate Lyase Activity | Li X | 2024 Apr 26 |
|
| 38634956 | Clinical relevance of glycosylation in triple negative breast cancer: a review | Chakraborty M | 2024 Apr |
|
| 37884654 | COG-imposed Golgi functional integrity determines the onset of dark-induced senescence | Choi HS | 2023 Oct 26 |
|
 G49112ZN
G49112ZN
 G85196CF
G85196CF
