Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE G25520XG
G25520XG
Summary
- GlyTouCan ID
-
G25520XG
- IUPAC Condensed
- Gal(b1-4)GlcNAc(b1-2)Man(a1-3)[GlcNAc(b1-4)][Gal(b1-4)GlcNAc(b1-2)Man(a1-6)]Man(b1-4)GlcNAc(b1-4)[Fuc(a1-6)]GlcNAc(b1-
- Motifs
- N-Glycan complex N-acetyllactosamine Type 2 LN
- Subsumption Level
- Fully-defined saccharide
- Monoisotopic Mass
- 1989.73
3D Structures
GlycoShape
Organisms
| Organisms | Evidence |
|---|---|
| Mus musculus (house mouse) | |
| Rattus norvegicus (Norway rat) | |
| Homo sapiens (human) | |
| Coturnix japonica (Japanese quail) | |
| unidentified influenza virus |
Taxonomic Hierarchy
root
cellular organisms [inference]
GlycoGene Database (GGDB)
KEGG BRITE Database
- KEGG ID
GALAXY
Core Protein
| UniProt ID | Protein Name | Reference | Source |
|---|---|---|---|
| P31994 | Low affinity immunoglobulin gamma Fc region receptor II-b | ||
| P35253 | Envelope glycoprotein | ||
| P41222 | Prostaglandin-H2 D-isomerase | ||
| P56817 | Beta-secretase 1 | ||
| P98158 | Low-density lipoprotein receptor-related protein 2 | ||
| Q70626 | Envelope glycoprotein gp160 | ||
| Q7SIB2 | Collagen alpha-1(IV) chain | ||
| Q7SIB3 | Collagen alpha-2(IV) chain (Fragment) | ||
| Q8BYI9 | Tenascin-R |
Sequence Descriptors
- GlycoCT
-
RES 1b:b-dglc-HEX-1:5 2s:n-acetyl 3b:b-dglc-HEX-1:5 4s:n-acetyl 5b:b-dman-HEX-1:5 6b:a-dman-HEX-1:5 7b:b-dglc-HEX-1:5 8s:n-acetyl 9b:b-dgal-HEX-1:5 10b:b-dglc-HEX-1:5 11s:n-acetyl 12b:a-dman-HEX-1:5 13b:b-dglc-HEX-1:5 14s:n-acetyl 15b:b-dgal-HEX-1:5 16b:a-lgal-HEX-1:5|6:d LIN 1:1d(2+1)2n 2:1o(4+1)3d 3:3d(2+1)4n 4:3o(4+1)5d 5:5o(3+1)6d 6:6o(2+1)7d 7:7d(2+1)8n 8:7o(4+1)9d 9:5o(4+1)10d 10:10d(2+1)11n 11:5o(6+1)12d 12:12o(2+1)13d 13:13d(2+1)14n 14:13o(4+1)15d 15:1o(6+1)16d
- WURCS
- WURCS=2.0/5,11,10/[a2122h-1b_1-5_2*NCC/3=O][a1122h-1b_1-5][a1122h-1a_1-5][a2112h-1b_1-5][a1221m-1a_1-5]/1-1-2-3-1-4-1-3-1-4-5/a4-b1_a6-k1_b4-c1_c3-d1_c4-g1_c6-h1_d2-e1_e4-f1_h2-i1_i4-j1
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 10211705 | Evidence for a site-specific fucosylation of N-linked oligosaccharide of immunoglobulin A1 from normal human serum | Tanaka A | 1998 Oct |
|
| 8870235 | The resolution of the neutral N-linked oligosaccharides of IgG by high pH anion-exchange chromatography | McGuire JM | 1996 Oct 04 |
|
| 8612632 | Comparative study of the sugar chains of alkaline phosphatases purified from rat liver and rat AH-130 hepatoma cells. Occurrence of fucosylated high-mannose-type and hybrid-type sugar chains | Endo T | 1996 Mar 01 |
|
| 8620897 | Detailed oligosaccharide structures of human integrin alpha 5 beta 1 analyzed by a three-dimensional mapping technique | Nakagawa H | 1996 Apr 01 |
|
| 7785764 | Identification of neutral and sialyl N-linked oligosaccharide structures from human serum glycoproteins using three kinds of high-performance liquid chromatography | Nakagawa H | 1995 Mar 20 |
|
| 8174273 | Alteration of asparagine-linked glycosylation in serum transferrin of patients with hepatocellular carcinoma | Matsumoto K | 1994 Jan 14 |
|
| 7525874 | Carbohydrate Structures of β‐Trace Protein from Human Cerebrospinal Fluid: Evidence for “Brain‐Type”N‐Glycosylation | Hoffmann A | 1994 Dec |
|
| 7944531 | Carbohydrate deficient glycoprotein syndrome type II: a deficiency in Golgi localised N-acetyl-glucosaminyltransferase II | Jaeken J | 1994 Aug |
|
| 8251489 | Structural study of the sugar chains of human leukocyte common antigen CD45 | Sato T | 1993 Nov 01 |
|
| 7679920 | Carbohydrate structures of human alpha-fetoprotein of patients with hepatocellular carcinoma: presence of fucosylated and non-fucosylated triantennary glycans | Aoyagi Y | 1993 Mar |
|
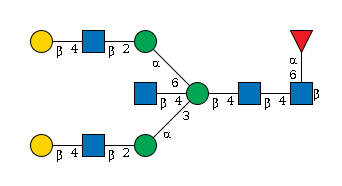
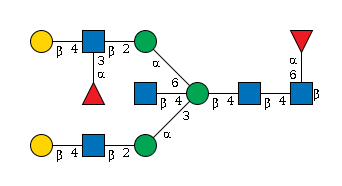 G95617FJ
G95617FJ
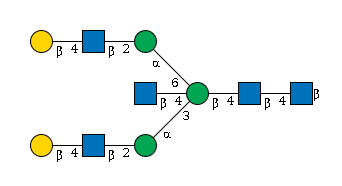 G52934AK
G52934AK
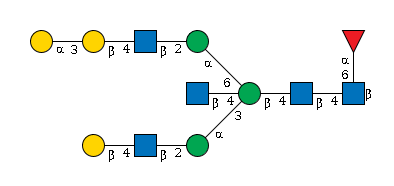 G61684ET
G61684ET
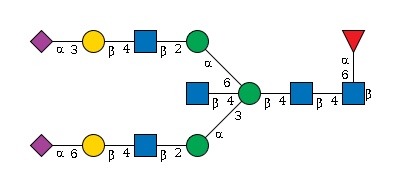 G94045IP
G94045IP
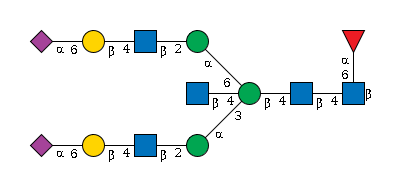 G14127XU
G14127XU
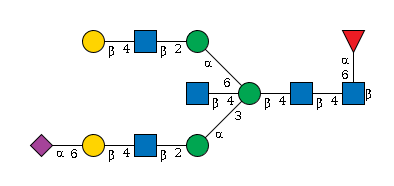 G91023GB
G91023GB
