Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE G00031MO
G00031MO
Summary
- GlyTouCan ID
-
G00031MO
- IUPAC Condensed
- Gal(b1-3)GalNAc(a1-
- Motifs
- O-Glycan core 1 T antigen TF antigen
- Subsumption Level
- Fully-defined saccharide
- Monoisotopic Mass
- 383.14
External Links
- GlyConnect structure
- GlyGen
- ChEBI
- PubChem Compound
- PubChem Substance
- CarbBank
-
- 98
- 99
- 100
- 2225
- 2226
- 2227
- 2228
- 2229
- 2230
- 2231
- 2232
- 2234
- 2235
- 2236
- 2237
- 2238
- 2239
- 2240
- 2241
- 2242
- 2243
- 3235
- 3236
- 3239
- 3240
- 3245
- 3255
- 3406
- 3795
- 3803
- 3842
- 3846
- 3903
- 4685
- 5035
- 5038
- 5203
- 5232
- 5434
- 5887
- 14321
- 20223
- 20981
- 25363
- 25729
- 25730
- 28182
- 29012
- 29046
- 29175
- 29393
- 29521
- 29709
- 30501
- 30734
- 30735
- 30848
- 30849
- 30850
- 30917
- 32646
- 33002
- 33022
- 33211
- 33469
- 33576
- 33832
- 33848
- 33851
- 33878
- 33952
- 34693
- 34766
- 34823
- 34829
- 34830
- 34986
- 34995
- 35029
- 35050
- 35069
- 35107
- 35108
- 35805
- 35833
- 35991
- 36264
- 36818
- 36826
- 36863
- 37676
- 37768
- 37770
- 38102
- 38263
- 38587
- 38640
- 38672
- 38687
- 40630
- 40825
- 40957
- 41263
- 41866
- 42208
- 42209
- 42740
- 42797
- 42875
- 43908
- 43915
- 43921
- 43923
- 43951
- 44029
- 44376
- 44505
- 44775
- 45972
- 45973
- 45974
- 45975
- 45976
- 45977
- 45978
- 45979
- 45980
- 45981
- 46056
- 46438
- 46466
- 46562
- 47015
- 47100
- 47186
- 47327
- 47600
- 47770
- 48244
- 48247
- 48891
- 49121
- 49283
- 49507
- 50477
- 50865
3D Structures
- GLYCAM
- DGalpb1-3DGalpNAca1-OH
Organisms
| Organisms | Evidence |
|---|---|
| Gadus | |
| Cricetulus griseus (Chinese hamster) | |
| Mus musculus (house mouse) | |
| Bos taurus (domestic cattle) | |
| Rattus |
Taxonomic Hierarchy
root
cellular organisms [inference]
Eukaryota (eucaryotes) [inference]
Opisthokonta [inference]
Metazoa (metazoans) [inference]
GlycoGene Database (GGDB)
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| C1GALT1 | (not applicable) |
 G57321FI
G57321FI
|
Ser/Thr |
 G00031MO
G00031MO
|
Ser/Thr | |
| ST6GALNAC1 | CMP-Neu5Ac |
 G00031MO
G00031MO
|
Ser/Thr |
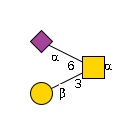 G56682BC
G56682BC
|
Ser/Thr | |
| ST3GAL2 | CMP-Neu5Ac |
 G00031MO
G00031MO
|
R |
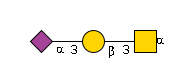 G65562ZE
G65562ZE
|
R | |
| ST6GALNAC2 | CMP-Neu5Ac |
 G00031MO
G00031MO
|
Ser/Thr |
 G56682BC
G56682BC
|
Ser/Thr | |
| ST3GAL4 | CMP-Neu5Ac |
 G00031MO
G00031MO
|
free |
 G65562ZE
G65562ZE
|
free |
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| C1GALT1 | (not applicable) |
 G57321FI
G57321FI
|
Ser/Thr |
 G00031MO
G00031MO
|
Ser/Thr | |
| ST3GAL3 | CMP-Neu5Ac |
 G00031MO
G00031MO
|
free |
 G65562ZE
G65562ZE
|
free | |
| ST6GALNAC1 | CMP-Neu5Ac |
 G00031MO
G00031MO
|
Ser/Thr |
 G56682BC
G56682BC
|
Ser/Thr | |
| ST3GAL2 | CMP-Neu5Ac |
 G00031MO
G00031MO
|
R |
 G65562ZE
G65562ZE
|
R | |
| ST6GALNAC1 | CMP-Neu5Ac |
 G00031MO
G00031MO
|
O-glycan Synthesis |
 G56682BC
G56682BC
|
O-glycan Synthesis |
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Reference |
|---|---|---|---|---|
| ST6GAL2 | CMP-Neu5Ac |
 G00031MO
G00031MO
|
free | |
| ST6GALNAC4 | CMP-Neu5Ac |
 G00031MO
G00031MO
|
benzyl | |
| ST6GALNAC3 | CMP-Neu5Ac |
 G00031MO
G00031MO
|
Ser/Thr | |
| ST3GAL3 | CMP-Neu5Ac |
 G00031MO
G00031MO
|
Antifreeze glycoprotein | |
| ST3GAL6 | CMP-Neu5Ac |
 G00031MO
G00031MO
|
free |
KEGG BRITE Database
- KEGG ID
- Enzyme
- References
- Reactions
Core Protein
| UniProt ID | Protein Name | Reference | Source |
|---|---|---|---|
| P01880 | Immunoglobulin heavy constant delta | ||
| P02668 | Kappa-casein | ||
| P02724 | Glycophorin-A | ||
| P02787 | Serotransferrin | ||
| P03393 | Glycoprotein 55 | ||
| P04275 | von Willebrand factor | ||
| P05059 | Chromogranin-A | ||
| P07314 | Glutathione hydrolase 1 proenzyme | ||
| P07498 | Kappa-casein | ||
| P08519 | Apolipoprotein(a) |
GlycoEpitope
- Epitope ID
- EP0020
Pathway
| Pathway Name | Organism |
|---|---|
| E.coli O107 | Escherichia coli |
| E.coli O117 | Escherichia coli |
| E.coli O185 | Escherichia coli |
| E.coli O5ab | Escherichia coli |
| E.coli O5ac | Escherichia coli |
| E.coli O70 | Escherichia coli |
| E.coli O71 | Escherichia coli |
| E.coli O83 | Escherichia coli |
Sequence Descriptors
- GlycoCT
-
RES 1b:a-dgal-HEX-1:5 2s:n-acetyl 3b:b-dgal-HEX-1:5 LIN 1:1d(2+1)2n 2:1o(3+1)3d
- WURCS
- WURCS=2.0/2,2,1/[a2112h-1a_1-5_2*NCC/3=O][a2112h-1b_1-5]/1-2/a3-b1
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 28096352 | Recognition of protein-linked glycans as a determinant of peptidase activity | Noach I | 2017 Jan 17 |
|
| 27667696 | Inflammation-Induced Adhesin-Receptor Interaction Provides a Fitness Advantage to Uropathogenic E. coli during Chronic Infection | Conover MS | 2016 Oct 12 |
|
| 27526028 | Structural basis of laminin binding to the LARGE glycans on dystroglycan | Briggs DC | 2016 Aug 15 |
|
| 26304114 | Structural Analysis of a Family 101 Glycoside Hydrolase in Complex with Carbohydrates Reveals Insights into Its Mechanism | Gregg KJ | 2015 Oct 16 |
|
| 25759508 | Hinge-Region O-Glycosylation of Human Immunoglobulin G3 (IgG3) | Plomp R | 2015 May |
|
| 26105055 | Structural Hot Spots Determine Functional Diversity of the Candida glabrata Epithelial Adhesin Family | Diderrich R | 2015 Aug 07 |
|
| 24915077 | Structures and binding specificity of galactose- and mannose-binding lectins from champedak: differences from jackfruit lectins | Gabrielsen M | 2014 May 24 |
|
| 25187573 | The O-glycomap of Lubricin, a Novel Mucin Responsible for Joint Lubrication, Identified by Site-specific Glycopeptide Analysis | Ali L | 2014 Dec |
|
| 23782692 | Recognition of the Thomsen-Friedenreich Pancarcinoma Carbohydrate Antigen by a Lamprey Variable Lymphocyte Receptor | Luo M | 2013 Jun 26 |
|
| 22678432 | Determination of site-specific glycan heterogeneity on glycoproteins | Kolarich D | 2012 Jun 07 |
|
PubAnnotation
| Title | Authors | Source | Publication Date | PubMed ID |
|---|---|---|---|---|
| The binding of simian virus 40 large T antigen to the polyphosphate backbone of nucleic acids. |
|
Biochim Biophys Acta | 1982 Jun 30 | 6896661 |
| Purification and characterization of the cytoplasmic casein kinase I from rat liver. |
|
Biochim Biophys Acta | 1982 Jan 18 | 6948579 |
| Use of monoclonal antibodies as probes of simian virus 40 T antigen ATPase activity. |
|
J Biol Chem | 1981 Nov 25 | 6170639 |
| Characteristics of an SV40-plasmid recombinant and its movement into and out of the genome of a murine cell. |
|
Cell | 1980 Aug | 6250708 |
| A stretch of "late" SV40 viral DNA about 400 bp long which includes the origin of replication is specifically exposed in SV40 minichromosomes. |
|
Cell | 1979 Feb | 222461 |
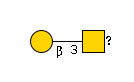 G00032MO
G00032MO
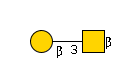 G01534TU
G01534TU
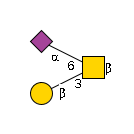 G28694FA
G28694FA
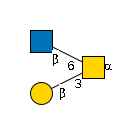 G00033MO
G00033MO
