Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE G00035MO
G00035MO
Summary
- GlyTouCan ID
-
G00035MO
- IUPAC Condensed
- GlcNAc(b1-3)GalNAc(a1-
- Motifs
- O-Glycan core 3
- Subsumption Level
- Fully-defined saccharide
- Monoisotopic Mass
- 424.17
3D Structures
- GLYCAM
- DGlcpNAcb1-3DGalpNAca1-OH
Organisms
| Organisms | Evidence |
|---|---|
| Rattus norvegicus (Norway rat) | |
| Homo sapiens (human) |
Taxonomic Hierarchy
GlycoGene Database (GGDB)
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| ST6GALNAC1 | CMP-Neu5Ac |
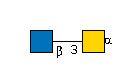 G00035MO
G00035MO
|
O-glycan Synthesis |
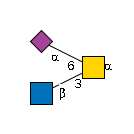 G63334FZ
G63334FZ
|
O-glycan Synthesis | |
| B4GALT4 | UDP-Gal |
 G00035MO
G00035MO
|
p-Nitrophenyl |
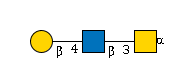 G32723SL
G32723SL
|
p-Nitrophenyl | |
| B3GALT5 | UDP-Gal |
 G00035MO
G00035MO
|
a-OpNp |
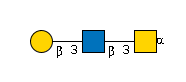 G11647UD
G11647UD
|
a-OpNp | |
| B4GALT1 | UDP-Gal |
 G00035MO
G00035MO
|
O-glycan Synthesis |
 G32723SL
G32723SL
|
O-glycan Synthesis | |
| B3GALT5 | UDP-Gal |
 G00035MO
G00035MO
|
O-glycan Synthesis |
 G11647UD
G11647UD
|
O-glycan Synthesis |
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Reference |
|---|---|---|---|---|
| C1GALT1 | UDP-Gal |
 G00035MO
G00035MO
|
O-glycan Synthesis | |
| B3GNT7 | UDP-GlcNAc |
 G00035MO
G00035MO
|
core 3-pNp | |
| A4GNT | UDP-GlcNAc |
 G00035MO
G00035MO
|
pNP | |
| B3GNT8 | UDP-GlcNAc |
 G00035MO
G00035MO
|
core 3-pNp | |
| GCNT4 | UDP-GlcNAc |
 G00035MO
G00035MO
|
[alpha]-1-p-nitrophenyl |
KEGG BRITE Database
Core Protein
Sequence Descriptors
- GlycoCT
-
RES 1b:a-dgal-HEX-1:5 2s:n-acetyl 3b:b-dglc-HEX-1:5 4s:n-acetyl LIN 1:1d(2+1)2n 2:1o(3+1)3d 3:3d(2+1)4n
- WURCS
- WURCS=2.0/2,2,1/[a2112h-1a_1-5_2*NCC/3=O][a2122h-1b_1-5_2*NCC/3=O]/1-2/a3-b1
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 34191522 | The O-Glycome of Human Nigrostriatal Tissue and Its Alteration in Parkinson's Disease | Wilkinson H | 2021 Aug 6 |
|
| 19422833 | N-Acetylglucosamine Recognition by a Family 32 Carbohydrate-Binding Module from Clostridium perfringens NagH | Ficko-Blean E | 2009 Jul 10 |
|
| 15620693 | A novel β1,3‐N‐acetylglucosaminyltransferase (β3Gn‐T8), which synthesizes poly‐N‐acetyllactosamine, is dramatically upregulated in colon cancer | Ishida H | 2004 Nov 28 |
|
| 15044014 | Molecular cloning and characterization of β1,4‐N‐acetylgalactosaminyltransferases IV synthesizing N,N′‐diacetyllactosediamine1 | Gotoh M | 2004 Mar 05 |
|
| 14724282 | A novel human beta1,3-N-acetylgalactosaminyltransferase that synthesizes a unique carbohydrate structure, GalNAcbeta1-3GlcNAc | Hiruma T | 2004 Apr 02 |
|
| 12966086 | Molecular cloning and characterization of a novel human beta 1,4-N-acetylgalactosaminyltransferase, beta 4GalNAc-T3, responsible for the synthesis of N,N'-diacetyllactosediamine, galNAc beta 1-4GlcNAc | Sato T | 2003 Nov 28 |
|
| 11821425 | Molecular cloning and characterization of a novel UDP-GlcNAc:GalNAc-peptide beta1,3-N-acetylglucosaminyltransferase (beta 3Gn-T6), an enzyme synthesizing the core 3 structure of O-glycans | Iwai T | 2002 Apr 12 |
|
| 10753916 | Control of O-glycan branch formation. Molecular cloning and characterization of a novel thymus-associated core 2 beta1, 6-n-acetylglucosaminyltransferase | Schwientek T | 2000 Apr 14 |
|
| 10406968 | Molecular cloning of a human UDP-galactose:GlcNAcbeta1,3GalNAc beta1, 3 galactosyltransferase gene encoding an O-linked core3-elongation enzyme | Zhou D | 1999 Jul 15 |
|
| 9915862 | Molecular cloning and expression of a novel beta-1, 6-N-acetylglucosaminyltransferase that forms core 2, core 4, and I branches | Yeh JC | 1999 Jan 29 |
|
PubAnnotation
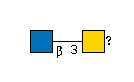 G00036MO
G00036MO
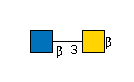 G75113PZ
G75113PZ
 G57321FI
G57321FI
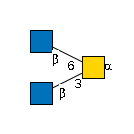 G00037MO
G00037MO
