Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE G57321FI
G57321FI
Summary
- GlyTouCan ID
-
G57321FI
- IUPAC Condensed
- GalNAc(a1-
- Motifs
- Tn antigen
Organisms
| Organisms | Evidence |
|---|---|
| Mesocestoides vogae | |
| Chlorocebus aethiops (grivet) | |
| Oryctolagus cuniculus (rabbit) | |
| Saccharomyces cerevisiae (brewer's yeast) | |
| Taenia hydatigena |
Taxonomic Hierarchy
root
cellular organisms [inference]
Eukaryota (eucaryotes) [inference]
Opisthokonta [inference]
Metazoa (metazoans) [inference]
GlycoGene Database (GGDB)
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| C1GALT1 | (not applicable) |
 G57321FI
G57321FI
|
Ser/Thr |
 G00031MO
G00031MO
|
Ser/Thr | |
| ST6GALNAC1 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
Ser/Thr |
 G36123IU
G36123IU
|
Ser/Thr | |
| ST6GALNAC1 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
O-glycan Synthesis |
 G36123IU
G36123IU
|
O-glycan Synthesis | |
| B3GNT6 | UDP-GlcNAc |
 G57321FI
G57321FI
|
[alpha]-pNP |
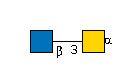 G00035MO
G00035MO
|
[alpha]-pNP |
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| C1GALT1 | (not applicable) |
 G57321FI
G57321FI
|
Ser/Thr |
 G00031MO
G00031MO
|
Ser/Thr | |
| ST6GALNAC1 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
Ser/Thr |
 G36123IU
G36123IU
|
Ser/Thr | |
| ST6GALNAC4 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
benzyl |
 G36123IU
G36123IU
|
benzyl | |
| ST6GALNAC1 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
O-glycan Synthesis |
 G36123IU
G36123IU
|
O-glycan Synthesis | |
| ST6GALNAC1 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
[Ala-Thr(*)-Ala]2-7 |
 G36123IU
G36123IU
|
[Ala-Thr(*)-Ala]2-7 |
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Reference |
|---|---|---|---|---|
| ST6GALNAC3 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
Ser/Thr | |
| ST6GAL2 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
para-nitrophenol | |
| ST6GALNAC2 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
benzyl | |
| ST6GALNAC1 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
[alpha]-Bz | |
| ST6GALNAC2 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
Ser/Thr |
KEGG BRITE Database
Core Protein
GlycoEpitope
- Epitope ID
- EP0021
Pathway
| Pathway Name | Organism |
|---|---|
| E.coli O86H2K2 | Escherichia coli |
| E.coli O86H2K62 | Escherichia coli |
| E.coli O90 | Escherichia coli |
| Ficolins bind to repetitive carbohydrate structures on the target cell surface | Xenopus tropicalis |
| Ficolins bind to repetitive carbohydrate structures on the target cell surface | Homo sapiens |
| Ficolins bind to repetitive carbohydrate structures on the target cell surface | Gallus gallus |
| Ficolins bind to repetitive carbohydrate structures on the target cell surface | Mus musculus |
| Ficolins bind to repetitive carbohydrate structures on the target cell surface | Rattus norvegicus |
| Ficolins bind to repetitive carbohydrate structures on the target cell surface | Bos taurus |
| Ficolins bind to repetitive carbohydrate structures on the target cell surface | Canis familiaris |
Sequence Descriptors
- GlycoCT
-
RES 1b:a-dgal-HEX-1:5 2s:n-acetyl LIN 1:1d(2+1)2n
- WURCS
- WURCS=2.0/1,1,0/[a2112h-1a_1-5_2*NCC/3=O]/1/
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 3998961 | Cuticular Carbohydrates of Three Nematode Species and Chemoreception by Trichostrongylus colubriformis | Bone LW | 1985 Apr |
|
| 3002784 | Validity of Lymphoid Cell Line for Enzymatic Studies of GM2-Gangliosidosis Variant 0 (Sandhoff Disease) | Maret A | 1985 |
|
| 3868975 | Structural studies on a sulphated glycoprotein preparation isolated from human saliva | Green DR | 1985 |
|
| 3928981 | B cell growth and differentiation activity of a purified T cell-replacing factor (TRF) molecule from B151-T cell hybridoma | Hara Y | 1985 |
|
| 6487322 | The B4 lectin from Vicia villosa seeds interacts with N-acetylgalactosamine residues on erythrocytes with blood group Cad specificity | Tollefsen SE | 1984 Sep 28 |
|
| 6547858 | Presence of two neutral disaccharides containing N-acetylhexosamine in bovine colostrum as free forms | Saito T | 1984 Sep 07 |
|
| 6466706 | Evidence for the occurence of oligosaccharides from bovine glycophorin having a branching point at C-6 of N-acetyl-galactosamine | Fukuda K | 1984 Sep 07 |
|
| 6471810 | Appearance of Helix pomatia lectin-binding sites on podocyte plasma membrane during glomerular differentiation. A quantitative analysis using the lectin-gold technique | Kunz A | 1984 Sep |
|
| 6335274 | [Quantification of lectin receptors on B, T, T-gamma, and T-mu lymphocytes. II. PHA, SBA, Dolichos biflorus, and Tetragonolobus purpureus] | de Dios I | 1984 Sep |
|
| 6727869 | Differential binding characteristics and applications of DGal beta 1----3DGalNAc specific lectins | Wu AM | 1984 Sep |
|
PubAnnotation
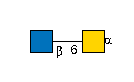 G00041MO
G00041MO
