Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE G57321FI
G57321FI
Summary
- GlyTouCan ID
-
G57321FI
- IUPAC Condensed
- GalNAc(a1-
- Motifs
- Tn antigen
Organisms
| Organisms | Evidence |
|---|---|
| Mesocestoides vogae | |
| Chlorocebus aethiops (grivet) | |
| Oryctolagus cuniculus (rabbit) | |
| Saccharomyces cerevisiae (brewer's yeast) | |
| Taenia hydatigena |
Taxonomic Hierarchy
root
cellular organisms [inference]
Eukaryota (eucaryotes) [inference]
Opisthokonta [inference]
Metazoa (metazoans) [inference]
GlycoGene Database (GGDB)
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| C1GALT1 | (not applicable) |
 G57321FI
G57321FI
|
Ser/Thr |
 G00031MO
G00031MO
|
Ser/Thr | |
| ST6GALNAC1 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
Ser/Thr |
 G36123IU
G36123IU
|
Ser/Thr | |
| ST6GALNAC1 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
O-glycan Synthesis |
 G36123IU
G36123IU
|
O-glycan Synthesis | |
| B3GNT6 | UDP-GlcNAc |
 G57321FI
G57321FI
|
[alpha]-pNP |
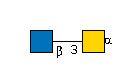 G00035MO
G00035MO
|
[alpha]-pNP |
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| C1GALT1 | (not applicable) |
 G57321FI
G57321FI
|
Ser/Thr |
 G00031MO
G00031MO
|
Ser/Thr | |
| ST6GALNAC1 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
Ser/Thr |
 G36123IU
G36123IU
|
Ser/Thr | |
| ST6GALNAC4 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
benzyl |
 G36123IU
G36123IU
|
benzyl | |
| ST6GALNAC1 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
O-glycan Synthesis |
 G36123IU
G36123IU
|
O-glycan Synthesis | |
| ST6GALNAC1 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
[Ala-Thr(*)-Ala]2-7 |
 G36123IU
G36123IU
|
[Ala-Thr(*)-Ala]2-7 |
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Reference |
|---|---|---|---|---|
| ST6GALNAC3 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
Ser/Thr | |
| ST6GAL2 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
para-nitrophenol | |
| ST6GALNAC2 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
benzyl | |
| ST6GALNAC1 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
[alpha]-Bz | |
| ST6GALNAC2 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
Ser/Thr |
KEGG BRITE Database
Core Protein
GlycoEpitope
- Epitope ID
- EP0021
Pathway
| Pathway Name | Organism |
|---|---|
| Ficolins bind to repetitive carbohydrate structures on the target cell surface | Sus scrofa |
| Initial triggering of complement | Mus musculus |
| Initial triggering of complement | Gallus gallus |
| Initial triggering of complement | Xenopus tropicalis |
| Initial triggering of complement | Sus scrofa |
| Initial triggering of complement | Bos taurus |
| Initial triggering of complement | Canis familiaris |
| Initial triggering of complement | Rattus norvegicus |
| Initial triggering of complement | Homo sapiens |
| Lectin pathway of complement activation | Bos taurus |
Sequence Descriptors
- GlycoCT
-
RES 1b:a-dgal-HEX-1:5 2s:n-acetyl LIN 1:1d(2+1)2n
- WURCS
- WURCS=2.0/1,1,0/[a2112h-1a_1-5_2*NCC/3=O]/1/
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 36293310 | Mutation Hotspot for Changing the Substrate Specificity of β-N-Acetylhexosaminidase: A Library of GlcNAcases | Nekvasilová P | 2022 Oct 18 |
|
| 36094307 | Disruption of the tagF Orthologue in the epa Locus Variable Region of Enterococcus faecalis Causes Cell Surface Changes and Suppresses an eep -Dependent Lysozyme Resistance Phenotype | Rouchon CN | 2022 Oct 18 |
|
| 35987426 | Differential expression of glycans in the urothelial layers of horse urinary bladder | Desantis S | 2022 Oct |
|
| 36253014 | Metabolomic markers of glucose regulation after a lifestyle intervention in prediabetes | Sevilla-Gonzalez MDR | 2022 Oct |
|
| 36041884 | Plasma and Liver Pharmacokinetics of the N-Acetylgalactosamine Short Interfering RNA JNJ-73763989 in Recombinant Adeno-Associated–Hepatitis B Virus–Infected Mice | Sandra L | 2022 Oct |
|
| 35907378 | Mn-doped bimetallic synergistic catalysis boosts for enzymatic phosphorylation of N-Acetylglucosamine/ N-Acetylgalactosamine and their derivatives | Xu H | 2022 Nov |
|
| 35618750 | Author Correction: The Pel polysaccharide is predominantly composed of a dimeric repeat of α-1,4 linked galactosamine and N-acetylgalactosamine | Le Mauff F | 2022 May 26 |
|
| 35504880 | Structural basis for the synthesis of the core 1 structure by C1GalT1 | González-Ramírez AM | 2022 May 03 |
|
| 35321541 | Protein Engineering of PhUGT, a Donor Promiscuous Glycosyltransferase, for the Improved Enzymatic Synthesis of Antioxidant Quercetin 3-O-N-Acetylgalactosamine | Xu Z | 2022 Mar 24 |
|
| 35151686 | Loss of the N-acetylgalactosamine side chain of the GPI-anchor impairs bone formation and brain functions and accelerates the prion disease pathology | Hirata T | 2022 Mar |
|
PubAnnotation
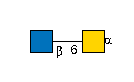 G00041MO
G00041MO
