Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE G57321FI
G57321FI
Summary
- GlyTouCan ID
-
G57321FI
- IUPAC Condensed
- GalNAc(a1-
- Motifs
- Tn antigen
Organisms
| Organisms | Evidence |
|---|---|
| Mesocestoides vogae | |
| Chlorocebus aethiops (grivet) | |
| Oryctolagus cuniculus (rabbit) | |
| Saccharomyces cerevisiae (brewer's yeast) | |
| Taenia hydatigena |
Taxonomic Hierarchy
root
cellular organisms [inference]
Eukaryota (eucaryotes) [inference]
Opisthokonta [inference]
Metazoa (metazoans) [inference]
GlycoGene Database (GGDB)
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| C1GALT1 | (not applicable) |
 G57321FI
G57321FI
|
Ser/Thr |
 G00031MO
G00031MO
|
Ser/Thr | |
| ST6GALNAC1 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
Ser/Thr |
 G36123IU
G36123IU
|
Ser/Thr | |
| ST6GALNAC1 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
O-glycan Synthesis |
 G36123IU
G36123IU
|
O-glycan Synthesis | |
| B3GNT6 | UDP-GlcNAc |
 G57321FI
G57321FI
|
[alpha]-pNP |
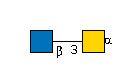 G00035MO
G00035MO
|
[alpha]-pNP |
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| C1GALT1 | (not applicable) |
 G57321FI
G57321FI
|
Ser/Thr |
 G00031MO
G00031MO
|
Ser/Thr | |
| ST6GALNAC1 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
Ser/Thr |
 G36123IU
G36123IU
|
Ser/Thr | |
| ST6GALNAC4 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
benzyl |
 G36123IU
G36123IU
|
benzyl | |
| ST6GALNAC1 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
O-glycan Synthesis |
 G36123IU
G36123IU
|
O-glycan Synthesis | |
| ST6GALNAC1 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
[Ala-Thr(*)-Ala]2-7 |
 G36123IU
G36123IU
|
[Ala-Thr(*)-Ala]2-7 |
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Reference |
|---|---|---|---|---|
| ST6GALNAC3 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
Ser/Thr | |
| ST6GAL2 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
para-nitrophenol | |
| ST6GALNAC2 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
benzyl | |
| ST6GALNAC1 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
[alpha]-Bz | |
| ST6GALNAC2 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
Ser/Thr |
KEGG BRITE Database
Core Protein
| UniProt ID | Protein Name | Reference | Source |
|---|---|---|---|
| O43660 | Pleiotropic regulator 1 | ||
| O43674 | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 5, mitochondrial | ||
| O43760 | Synaptogyrin-2 | ||
| O43768 | Alpha-endosulfine | ||
| O43852 | Calumenin | ||
| O43854 | EGF-like repeat and discoidin I-like domain-containing protein 3 | ||
| O43909 | Exostosin-like 3 | ||
| O60279 | Sushi domain-containing protein 5 | ||
| O60309 | Leucine-rich repeat-containing protein 37A3 | ||
| O60353 | Frizzled-6 |
GlycoEpitope
- Epitope ID
- EP0021
Pathway
| Pathway Name | Organism |
|---|---|
| E.coli O101 | Escherichia coli |
| E.coli O103 | Escherichia coli |
| E.coli O107 | Escherichia coli |
| E.coli O112ab | Escherichia coli |
| E.coli O112ac | Escherichia coli |
| E.coli O116 | Escherichia coli |
| E.coli O117 | Escherichia coli |
| E.coli O125ab | Escherichia coli |
| E.coli O127 | Escherichia coli |
| E.coli O128ab-H27 | Escherichia coli |
Sequence Descriptors
- GlycoCT
-
RES 1b:a-dgal-HEX-1:5 2s:n-acetyl LIN 1:1d(2+1)2n
- WURCS
- WURCS=2.0/1,1,0/[a2112h-1a_1-5_2*NCC/3=O]/1/
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 3094936 | The effect of penultimate N-acetylgalactosamine 4-sulfate on Chondroitin chain elongation | Cogburn JN | 1986 Aug 15 |
|
| 3768888 | Isolation and characterization of a receptor lectin specific for galactose/N-acetylgalactosamine from macrophages | Kawasaki T | 1986 Aug 15 |
|
| 3007660 | Characterization of a Herpes Simplex Virus Type 2-specified Glycoprotein with Affinity for N-Acetylgalactosamine-specific Lectins and Its Identification as g92K or gG | Olofsson S | 1986 Apr 01 |
|
| 3729878 | Lectin Binding and the Carbohydrate Moieties Present on Newcastle Disease Virus Strains | McMillan BC | 1986 Apr |
|
| 3702705 | Use of avidin-iminobiotin complexes for purifying plasma membrane proteins | Zeheb R | 1986 |
|
| 3274053 | Specificity of hemagglutinin of Falcata japonica which reacts with blood group active N-acetyl-D-galactosamine residues | Nakajima T | 1986 |
|
| 3757884 | [Localization of lectin receptors on the surface of C1300 neuroblastoma cells] | Khomutovskiĭ OA | 1986 |
|
| 2884962 | [Inhibition by antibodies of the adhesion of trophozoites of Entamoeba histolytica] | Barranco-Tovar C | 1986 |
|
| 2996612 | Galactose-specific receptors on liver cells. I. Hepatocyte and liver macrophage receptors differ in their membrane anchorage | Kempka G | 1985 Oct 30 |
|
| 4055733 | Sulfation of p-nitrophenyl-N-acetyl-beta-D-galactosaminide with a microsomal fraction from cultured chondrocytes. | Habuchi O | 1985 Oct 25 |
|
PubAnnotation
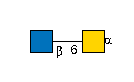 G00041MO
G00041MO
