Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE G80858MF
G80858MF
Summary
- GlyTouCan ID
-
G80858MF
- IUPAC Condensed
- GlcNAc(b1-2)Man(a1-3)[GlcNAc(b1-2)Man(a1-6)]Man(b1-4)GlcNAc(b1-4)[Fuc(a1-6)]GlcNAc(b1-
- Motifs
- N-Glycan complex
- Subsumption Level
- Fully-defined saccharide
- Monoisotopic Mass
- 1462.54
3D Structures
Organisms
| Organisms | Evidence |
|---|---|
| Cavia porcellus (domestic guinea pig) | |
| Chlorocebus aethiops (grivet) |
Taxonomic Hierarchy
root
cellular organisms [inference]
GlycoGene Database (GGDB)
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| FUT8 | GDP-Fuc |
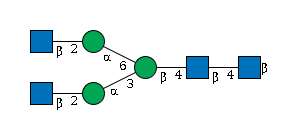 G39213VZ
G39213VZ
|
Free |
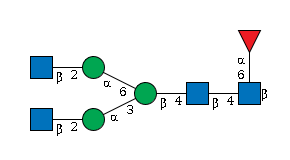 G80858MF
G80858MF
|
Free | |
| MGAT4B | UDP-GlcNAc |
 G80858MF
G80858MF
|
PA |
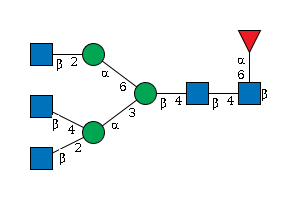 G61207RZ
G61207RZ
|
PA | |
| MGAT4A | UDP-GlcNAc |
 G80858MF
G80858MF
|
PA |
 G61207RZ
G61207RZ
|
PA | |
| MGAT4A | UDP-GlcNAc |
 G80858MF
G80858MF
|
Asn-X |
 G61207RZ
G61207RZ
|
Asn-X | |
| MGAT3 | UDP-GlcNAc |
 G80858MF
G80858MF
|
Asn-X |
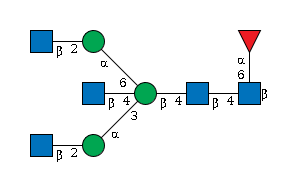 G30159WR
G30159WR
|
Asn-X |
KEGG BRITE Database
GALAXY
Core Protein
| UniProt ID | Protein Name | Reference | Source |
|---|---|---|---|
| O15389 | Sialic acid-binding Ig-like lectin 5 | ||
| O75015 | Low affinity immunoglobulin gamma Fc region receptor III-B | ||
| P00451 | Coagulation factor VIII | ||
| P00747 | Plasminogen | ||
| P01579 | Interferon gamma | ||
| P01854 | Immunoglobulin heavy constant epsilon | ||
| P01857 | Immunoglobulin heavy constant gamma 1 | ||
| P01859 | Immunoglobulin heavy constant gamma 2 | ||
| P01860 | Immunoglobulin heavy constant gamma 3 | ||
| P01861 | Immunoglobulin heavy constant gamma 4 |
Sequence Descriptors
- GlycoCT
-
RES 1b:b-dglc-HEX-1:5 2s:n-acetyl 3b:b-dglc-HEX-1:5 4s:n-acetyl 5b:b-dman-HEX-1:5 6b:a-dman-HEX-1:5 7b:b-dglc-HEX-1:5 8s:n-acetyl 9b:a-dman-HEX-1:5 10b:b-dglc-HEX-1:5 11s:n-acetyl 12b:a-lgal-HEX-1:5|6:d LIN 1:1d(2+1)2n 2:1o(4+1)3d 3:3d(2+1)4n 4:3o(4+1)5d 5:5o(3+1)6d 6:6o(2+1)7d 7:7d(2+1)8n 8:5o(6+1)9d 9:9o(2+1)10d 10:10d(2+1)11n 11:1o(6+1)12d
- WURCS
- WURCS=2.0/4,8,7/[a2122h-1b_1-5_2*NCC/3=O][a1122h-1b_1-5][a1122h-1a_1-5][a1221m-1a_1-5]/1-1-2-3-1-3-1-4/a4-b1_a6-h1_b4-c1_c3-d1_c6-f1_d2-e1_f2-g1
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 33242084 | Post-natal developmental changes in the composition of the rat neocortical N-glycome | Klarić Thomas S | 2021-06-03 |
|
| 33737529 | Toward robust N-glycomics of various tissue samples that may contain glycans with unknown or unexpected structures | Suzuki Noriko | 2021-03-18 |
|
| 34834161 | Glycan Profile Analysis of Engineered Trastuzumab with Rationally Added Glycosylation Sequons Presents Significantly Increased Glycan Complexity | Cruz E | 2021 Oct 20 |
|
| 33383989 | Bisecting GlcNAc Protein N-Glycosylation Is Characteristic of Human Adipogenesis | Wongtrakul-Kish K | 2021 Feb 5 |
|
| 34106099 | N-Glycosylation in isolated rat nerve terminals | Matthies I | 2021 Aug 1 |
|
| 31889402 | Glycan biomarkers for Alzheimer disease correlate with T-tau and P-tau in cerebrospinal fluid in subjective cognitive impairment | Schedin-Weiss Sophia | 2020-08 |
|
| 32841605 | Virus-Receptor Interactions of Glycosylated SARS-CoV-2 Spike and Human ACE2 Receptor | Zhao P | 2020 Oct 07 |
|
| 30674035 | Exploring Cerebrospinal Fluid IgG N-Glycosylation as Potential Biomarker for Amyotrophic Lateral Sclerosis | Costa J | 2019 Jan 23 |
|
| 29671580 | Site-Specific Profiling of Serum Glycoproteins Using N-Linked Glycan and Glycosite Analysis Revealing Atypical N-Glycosylation Sites on Albumin and alpha-1B-Glycoprotein | Sun S | 2018 Apr 19 |
|
| 26455811 | CSF N-glycoproteomics for early diagnosis in Alzheimer's disease | Palmigiano Angelo | 2016-01-10 |
|
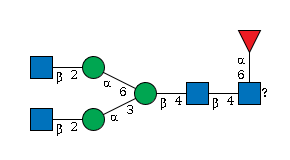 G89637WE
G89637WE
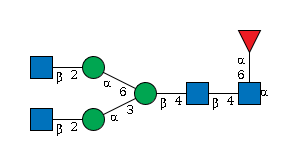 G95238UV
G95238UV
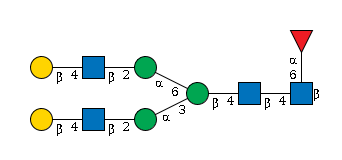 G78059CC
G78059CC
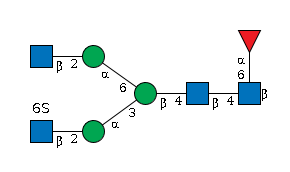 G59357BZ
G59357BZ
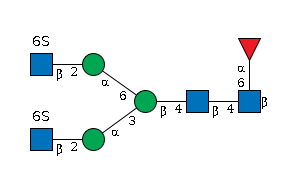 G65458HJ
G65458HJ
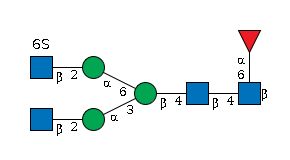 G54219FG
G54219FG
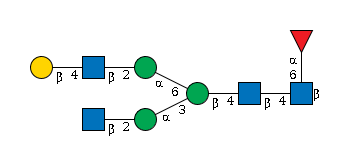 G27919IH
G27919IH
