Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE G36191CD
G36191CD
Summary
- GlyTouCan ID
-
G36191CD
- IUPAC Condensed
- Gal(b1-4)GlcNAc(b1-2)Man(a1-3)[Gal(b1-4)GlcNAc(b1-2)Man(a1-6)]Man(b1-4)GlcNAc(b1-4)GlcNAc(b1-
- Motifs
- N-Glycan complex N-acetyllactosamine Type 2 LN
- Subsumption Level
- Fully-defined saccharide
- Monoisotopic Mass
- 1640.59
External Links
- GlyConnect structure
- GlyGen
- ChEBI
- PubChem Compound
- PubChem Substance
- CarbBank
- 1608 1609 1610 2846 2848 2851 4374 4376 4377 4378 4628 4949 5082 5083 5412 6557 6558 6766 7427 7448 7705 7706 7708 7709 7710 7711 7712 7713 7714 7715 7716 7717 7985 8742 12767 12786 13312 14710 16631 17131 20303 20756 20757 20758 20883 25057 25744 29219 29848 30805 30826 31874 32380 33537 34452 34638 35753 35855 36436 37745 38434 40660 40669 42693 43551 43552 45811 46602 46815
3D Structures
Organisms
| Organisms | Evidence |
|---|---|
| Mus musculus (house mouse) | |
| Semliki Forest virus | |
| Cavia porcellus (domestic guinea pig) | |
| Coturnix japonica (Japanese quail) | |
| Rattus norvegicus (Norway rat) |
Taxonomic Hierarchy
root
cellular organisms [inference]
GlycoGene Database (GGDB)
KEGG BRITE Database
- KEGG ID
GALAXY
Core Protein
| UniProt ID | Protein Name | Reference | Source |
|---|---|---|---|
| P00742 | Coagulation factor X | ||
| P00747 | Plasminogen | ||
| P00797 | Renin | ||
| P01009 | Alpha-1-antitrypsin | ||
| P01215 | Glycoprotein hormones alpha chain | ||
| P01579 | Interferon gamma | ||
| P01588 | Erythropoietin | ||
| P01591 | Immunoglobulin J chain | ||
| P01730 | T-cell surface glycoprotein CD4 | ||
| P01854 | Immunoglobulin heavy constant epsilon |
Sequence Descriptors
- GlycoCT
-
RES 1b:b-dglc-HEX-1:5 2s:n-acetyl 3b:b-dglc-HEX-1:5 4s:n-acetyl 5b:b-dman-HEX-1:5 6b:a-dman-HEX-1:5 7b:b-dglc-HEX-1:5 8s:n-acetyl 9b:b-dgal-HEX-1:5 10b:a-dman-HEX-1:5 11b:b-dglc-HEX-1:5 12s:n-acetyl 13b:b-dgal-HEX-1:5 LIN 1:1d(2+1)2n 2:1o(4+1)3d 3:3d(2+1)4n 4:3o(4+1)5d 5:5o(3+1)6d 6:6o(2+1)7d 7:7d(2+1)8n 8:7o(4+1)9d 9:5o(6+1)10d 10:10o(2+1)11d 11:11d(2+1)12n 12:11o(4+1)13d
- WURCS
- WURCS=2.0/4,9,8/[a2122h-1b_1-5_2*NCC/3=O][a1122h-1b_1-5][a1122h-1a_1-5][a2112h-1b_1-5]/1-1-2-3-1-4-3-1-4/a4-b1_b4-c1_c3-d1_c6-g1_d2-e1_e4-f1_g2-h1_h4-i1
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 33242084 | Post-natal developmental changes in the composition of the rat neocortical N-glycome | Klarić Thomas S | 2021-06-03 |
|
| 33737529 | Toward robust N-glycomics of various tissue samples that may contain glycans with unknown or unexpected structures | Suzuki Noriko | 2021-03-18 |
|
| 34327564 | Identification of N-glycans with GalNAc-containing antennae from recombinant HIV trimers by ion mobility and negative ion fragmentation | Harvey DJ | 2021 Jul 30 |
|
| 33383989 | Bisecting GlcNAc Protein N-Glycosylation Is Characteristic of Human Adipogenesis | Wongtrakul-Kish K | 2021 Feb 5 |
|
| 29671580 | Site-Specific Profiling of Serum Glycoproteins Using N-Linked Glycan and Glycosite Analysis Revealing Atypical N-Glycosylation Sites on Albumin and alpha-1B-Glycoprotein | Sun S | 2018 Apr 19 |
|
| 25802287 | A Microarray-Matrix-assisted Laser Desorption/Ionization-Mass Spectrometry Approach for Site-specific Protein N-glycosylation Analysis, as Demonstrated for Human Serum Immunoglobulin M (IgM) | Pabst M | 2015 Jun |
|
| 21609021 | Mass Spectrometric Determination of IgG Subclass-Specific Glycosylation Profiles in Siblings Discordant for Myositis Syndromes | Perdivara I | 2011 Jul 1 |
|
| 20578731 | Identification of N-glycosylation changes in the CSF and serum in patients with schizophrenia | Stanta Johannes L | 2010-09-03 |
|
| 20131323 | Identification of N-glycans from Ebola virus glycoproteins by matrix-assisted laser desorption/ionisation time-of-flight and negative ion electrospray tandem mass spectrometry | Ritchie G | 2010 Feb 03 |
|
| 19088067 | Activity, splice variants, conserved peptide motifs, and phylogeny of two new alpha1,3-fucosyltransferase families (FUT10 and FUT11) | Mollicone R | 2009 Feb 13 |
|
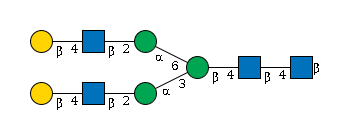
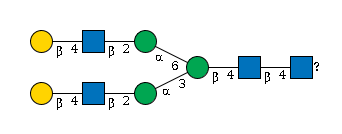 G61706MD
G61706MD
 G12201VG
G12201VG
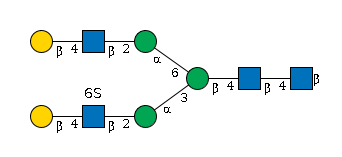 G09354YM
G09354YM
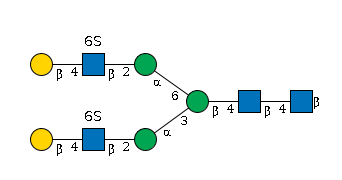 G45408HY
G45408HY
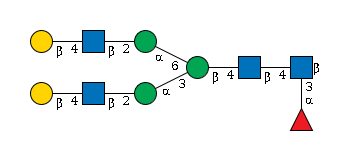 G27802LV
G27802LV
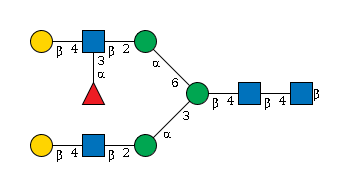 G01935FK
G01935FK
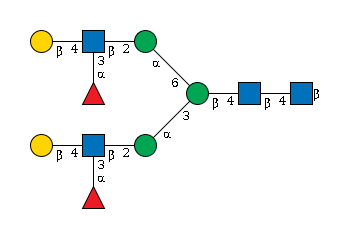 G25407RZ
G25407RZ
